Figure 3. Consensus m6A editing sites mapped to the NL4-3 genome.
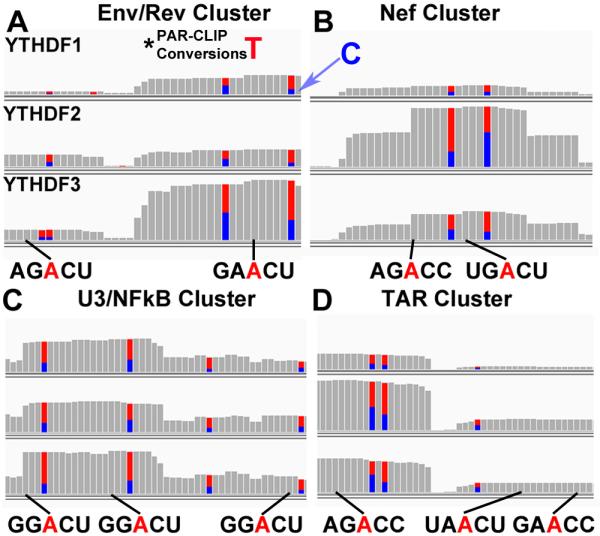
Shown in (A-D) are the 4 mapped YTHDF PAR-CLIP clusters present in NL4-3 with consensus m6A sites indicated. Adjacent T to C conversions, that result from 4SU photo-crosslinking (T=blue, C=red), are indicated. Below are the potential viral m6A editing sites shown in red, with a black line indicating the nucleotide position in the YTHDF binding cluster relative to the mutated T residue. This figure identifies all sites with a minimal (5’-RAC-3’) m6A consensus but this does not demonstrate that all of these A residues are actually modified. For related data, see Fig. S4.
