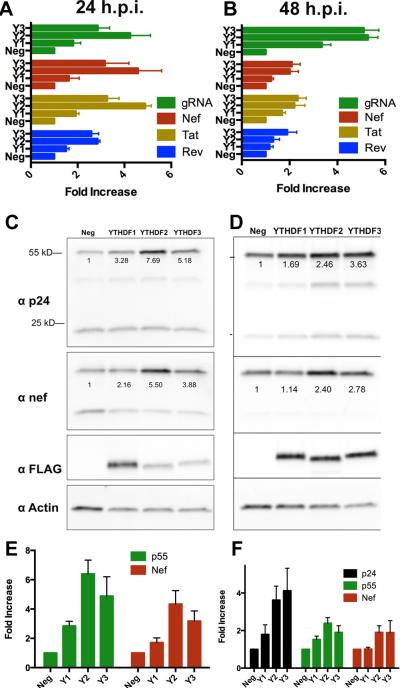
Figure 5. Overexpression of YTHDF m6A reader proteins boosts HIV-1 protein and RNA expression.
(A and B) qRT-PCR was used to quantify the expression level of the dominant spliced HIV-1 mRNA isoforms encoding Rev, Tat or Nef as well as the unspliced genomic RNA (gRNA). Assays were performed at 24 h (A) or 48 h (B) post-infection (hpi) using 293T cells stably overexpressing GFP (Neg) or one of the three YTHDF proteins (Y1 is YTHDF1 etc). Data were normalized to endogenous GAPDH mRNA. (C and D) Shown are representative Western blots from HIV-1 infection experiments similar to those described in (A and B). Infected 293T cells over-expressing GFP (Neg) or one of the YTHDF proteins were lysed at 24 hpi or 48 hpi then probed with an antibody specific for the HIV-1 p24 capsid protein, Nef, the FLAG tag on the overexpressed YTHDF protein or endogenous β-actin. Shown below the respective bands are actin-normalized quantifications. p55 represents uncleaved HIV-1 Gag polyprotein while p24 is the mature viral capsid (E and F). Shown are quantifications of band intensities from three independent Western experiments, similar to those shown in (C and D), performed at 24 hpi (E) or 48 hpi (F), with SD indicated.