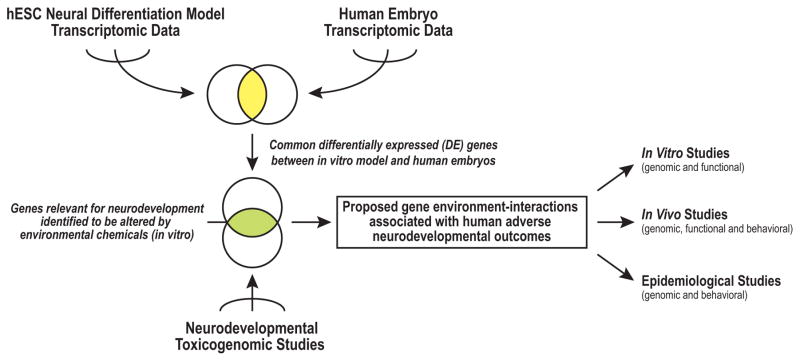
Figure 1. An analysis framework for integrating human transcriptomic datasets to identify biomarkers of neurodevelopmental toxicity.
Meta-analyses of transcriptomic data generated in human embryonic stem cell (hESC) neural differentiation models revealed signatures at the various stages, which were compared to datasets that interrogated gene expression at specific days of human embryonic development, including neurogenesis. Differentially expressed (DE) genes that were shared between the two models were chosen as candidate biomarkers of neural development. They were applied to toxicogenomic studies that employed hESCs to identify potential gene-environment interactions associated with adverse neurodevelopmental outcomes. These results could inform the design of future studies that use in vitro and in vivo models or investigate effects at a population level.