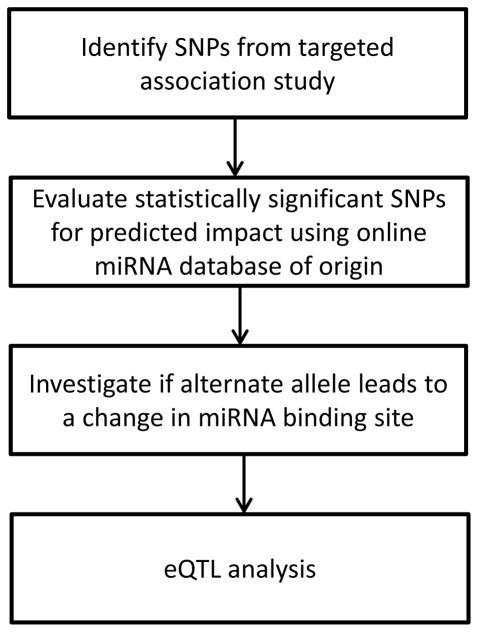
Figure 3.
Bioinformatic pipeline for predicted impact of statistically significant SNPs on miRNA target seed sites and cis gene expression. SNPs identified from the targeted association study were evaluated using PolymiRTS, Patrocles, and microRNA.org (in conjunction with markers identified by the 1000 Genomes Project) for indications of target seed sites that would be disrupted by the presence of the alternate allele for each genetic variant. Markers from the PolymiRTS database were further annotated for potential creation of new seed sites. Statistically significant markers were further investigated with MicroSNiPER to identify additional target seed sites both disrupted and potentially created by the presence of the alternate allele. The final step was cis-eQTL analysis where gene expression data was available.