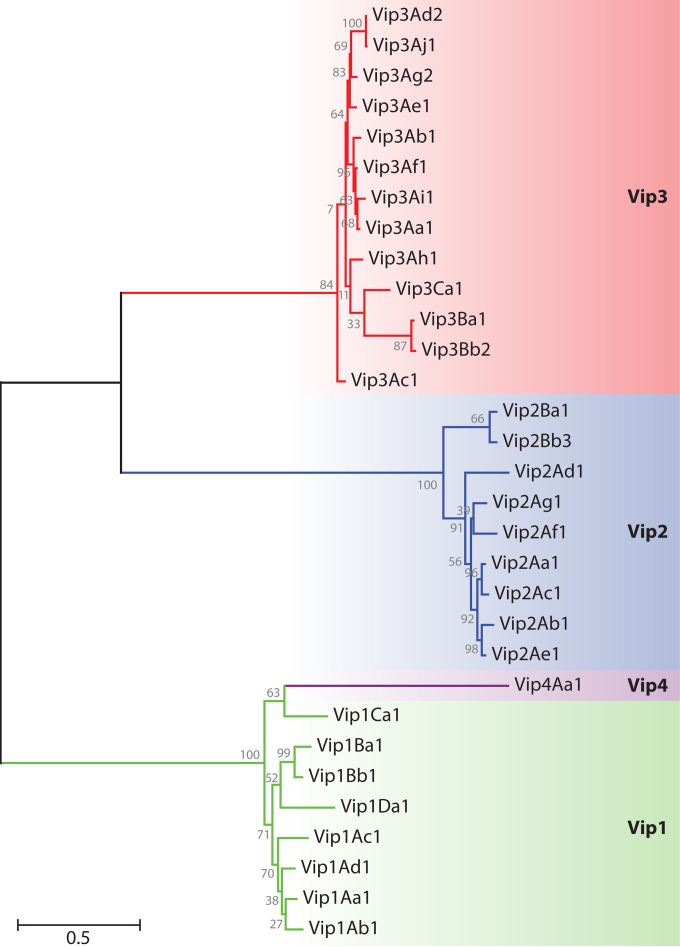
FIG 2.
Dendrogram showing the relationships among Vip proteins based on their degree of amino acid identity. Amino acid sequences were aligned by using the Clustal X interface (120). The evolutionary distance was calculated by maximum likelihood analysis, and the tree was constructed by using the MEGA5 program (121). The proteins used in this analysis are as follows: Vip1Aa1 (sequence identification number [Seq. ID no.] 5 in reference 28), Vip1Ab1 (Seq. ID no. 21 in reference 28), Vip1Ac1 (GenBank accession number HM439098), Vip1Ad1 (accession number JQ855505), Vip1Ba1 (accession number AAR40886), Vip1Bb1 (accession number AAR40282), Vip1Ca1 (accession number AAO86514), Vip1Da1 (accession number CAI40767), Vip2Aa1 (RCSB Protein Data Bank accession number 1QS1_A), Vip2Ab1 (Seq. ID no. 20 in reference 28), Vip2Ac1 (accession number AAO86513), Vip2Ad1 (accession number CAI40768), Vip2Ae1 (accession number EF442245), Vip2Af1 (accession number ACH42759), Vip2Ag1 (accession number JQ855506), Vip2Ba1 (accession number AAR40887), Vip2Bb3 (accession number AIA96500), Vip3Aa1 (accession number AAC37036), Vip3Ab1 (accession number AAR40284), Vip3Ac1 (named PS49C; Seq. ID no. 7 in K. Narva and D. Merlo, U.S. patent application 20,040,128,716), Vip3Ad2 (accession number CAI43276), Vip3Ae1 (accession number CAI43277), Vip3Af1 (accession number CAI43275), Vip3Ag2 (accession number ACL97352), Vip3Ah1 (accession number ABH10614), Vip3Ai1 (accession number KC156693), Vip3Aj1 (accession number KF826717), Vip3Ba1 (accession number AAV70653), Vip3Bb2 (accession number ABO30520), Vip3Ca1 (accession number ADZ46178), and Vip4Aa1 (accession number HM044666).