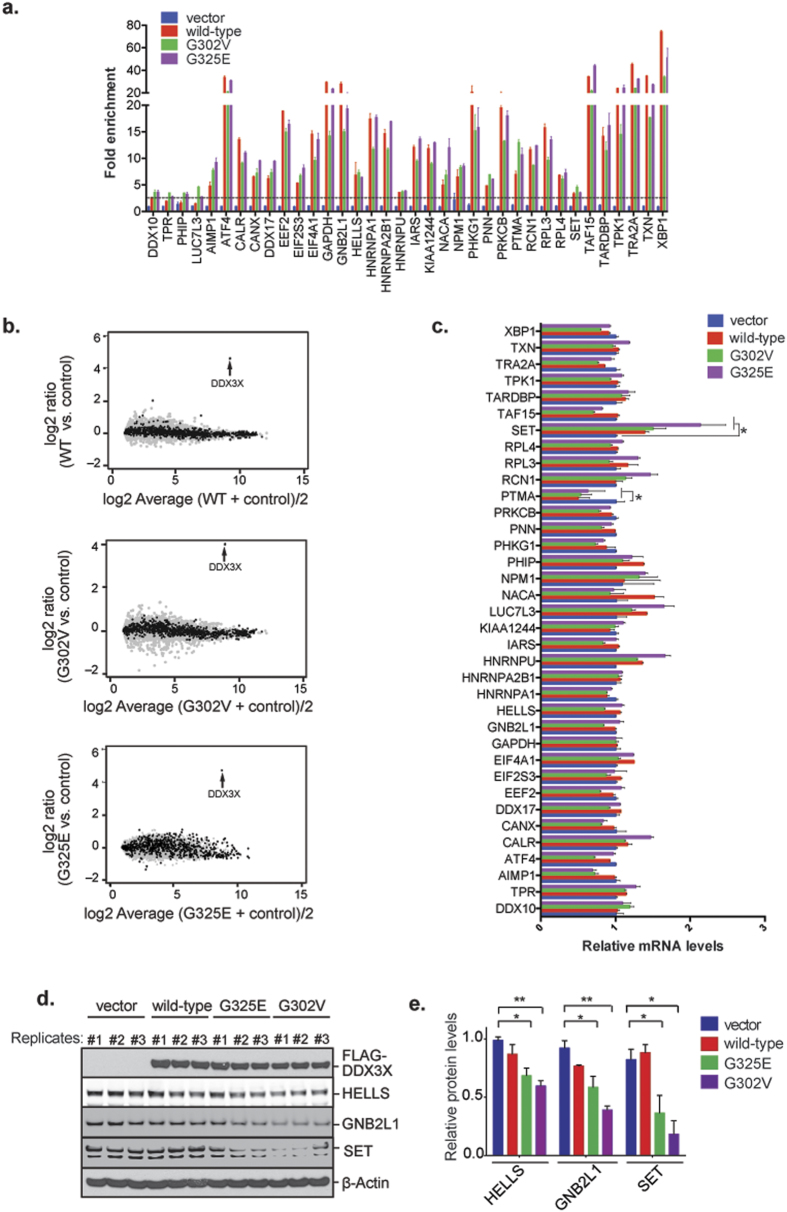
Figure 5. MB-associated mutations in DDX3X impact the translation of mRNA targets.
(a) RNA-immunoprecipitation analysis of numerous DDX3X targets using exogenous FLAG-tagged wild-type DDX3X and two cancer mutants (G302V and G325E). Two non-target RNAs were analyzed as negative control (DDX10 and TPR). Cells transfected with empty vector and immunoprecipitated with the anti-FLAG antibody M2 served as control. 87.5% of DDX3X mRNA targets were validated by the assay (horizontal black-dotted line represents cut-off based on non-target RNAs). (b) MA plot comparing RNA-seq data sets from HEK293T cells transfected with empty vector or a DDX3X-expressing vector (wild-type and two mutants: G302V and G325E). Transfections efficiencies are confirmed by the detection of high mRNA levels of DDX3X itself (denoted by the black arrows). Black dots represent DDX3X targets, gray dots represent non-targets. (c) Reverse transcription–qPCR analysis of several DDX3X CLIP-seq targets plus two non-targets (DDX10 and TPR) in HEK293T cells in cells expressing either wild-type DDX3X or two cancer-related mutant (G302V and G325E). Mean ± SEM values are based on a minimum of two replicated experiments. ~91% of DDX3X mRNA targets showed insignificant changes in their levels between cells expressing vector and those expressing DDX3X variants (Multifactorial ANOVA: *P ≤ 0.01). (d) Western blot analyses against FLAG and CLIP-seq targets HELLS, GNB2L1, and SET in HEK293T cells transfected with FLAG-tagged wild-type DDX3X or two cancer-related mutants (G325E and M370R). (e) The mean protein levels of three biological replicates of samples shown in (d) was graphed. Error bars represent mean values ± SEM (Student’s t-test between control or cells expressing DDX3X variants; *P ≤ 0.05; **P ≤ 0.01).