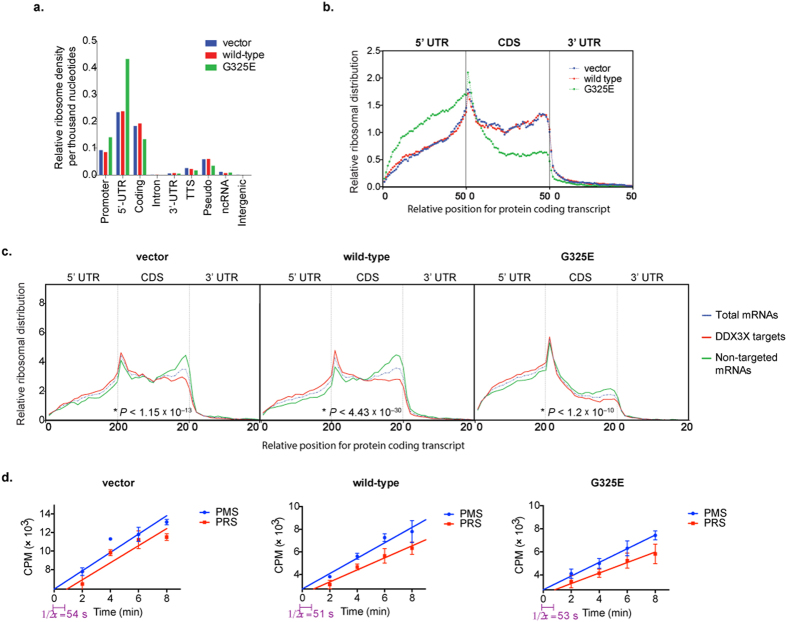
Figure 6. Ribosome profiling illustrates that expression of MB-associated DDX3X mutant G325E results in accumulation of ribosomes at 5′-UTR of mRNAs and impairs global translation.
(a) Ribosomal density per thousand nucleotides uniquely mapped to various annotated genomic regions (TTS, transcription termination sites; Pseudo, pseudogenes; ncRNA, non-coding RNAs; 5′-UTR, 5′- untranslated regions; 3′-UTR, 3′ untranslated regions; CDS, coding sequences). (b) Metagene analyses of ribosomal densities across the mRNA structure. Shown are the relative ribosomal density curves calculated for each of the 50-binned positions among three regions of the mRNA (5′-UTR, CDS, and 3′-UTR). Vertical lines separate the three regions of the mRNA. (c) MB-associated DDX3X mutant G325E impairs global translation. Metagene analyses of ribosomal densities across the mRNA structure for total mRNA (dashed-blue line), mRNAs identified as DDX3X targets (red lines), and mRNAs not identified as DDX3X targets (green lines) by CLIP-seq. The analysis was done in control cells or cells expressing wild-type DDX3X or the G325E mutant form. Shown are the relative ribosomal density curves calculated for each of the 20-binned positions among three regions of the mRNA (5′-UTR, CDS, and 3′-UTR). Vertical lines separate the three regions of the mRNA. The P-values for significant changes of ribosomal distribution between target or non-target mRNAs in the coding sequence was calculated using Kolmogorov-Smirnov equality-of-distribution test. (d) Ribosome half-transit analyses for cells expressing vector or FLAG-tagged wild-type DDX3X or MB-associated mutant DDX3X-G325E for 24 h in HEK293T cells. 35S-Met/Cys labeling incorporation into all polypeptides (postmitochondrial supernatant, PMS) and into polypeptide released from ribosomes (postribosomal supernatant, PRS) was obtained by linear regression analysis. Mean ± SEM values are based on a minimum of four replicated experiments. Ribosome half-transit time (1/2τ) were relatively equal in all samples.