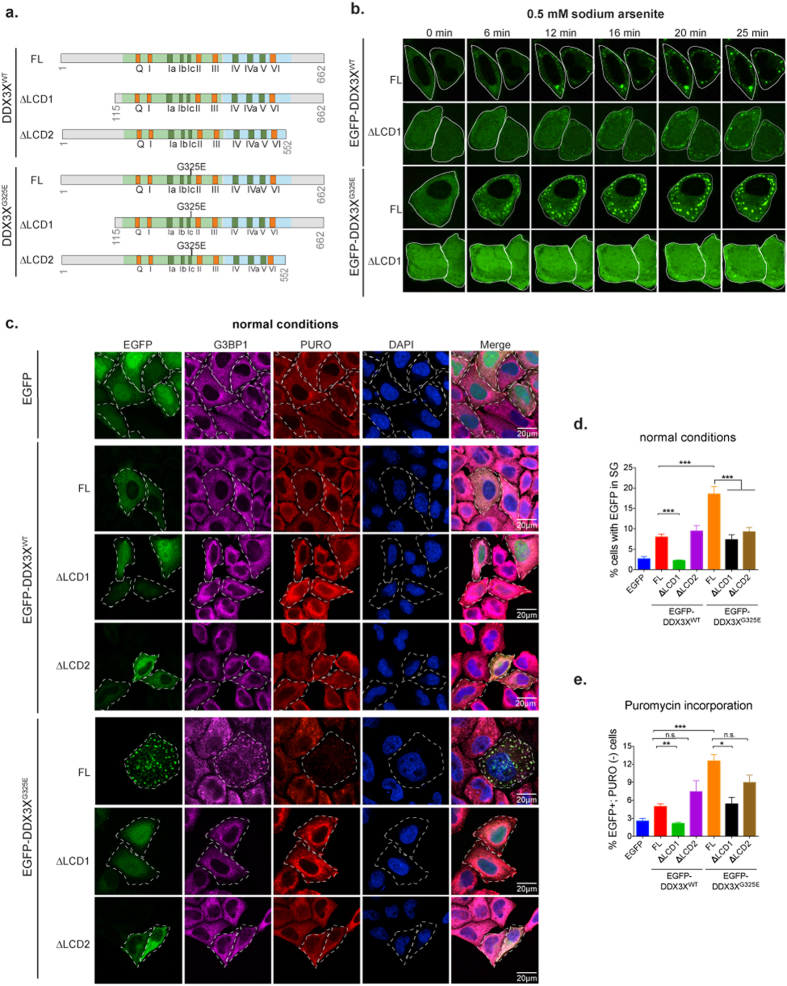
Figure 7. Deletion of N-terminal low complexity domain prevents cancer-related DDX3X mutant from inducing SG assembly and repressing mRNA translation.
(a) Schematic representation of full-length DDX3X or DDX3X with deletion of either N-terminal low complexity domain lacking amino acids 1–114 (ΔLCD1) or C-terminal low complexity domain lacking amino acids 553–662 (ΔLCD2). The ATPase/helicase and RNA binding functional motifs are denoted in orange and dark green boxes. The location of the MB-associated mutant G325E is shown in the scheme. (b) Live imaging of HeLa cells transfected with the indicated EGFP-tagged DDX3X constructs for 24 h and treated with sodium arsenite for a period of 25 min. Images were collected every 30 seconds. Representative images of the indicated time-points are shown. (c) Puromycin incorporation assay in HeLa cells transfected with EGFP alone or the indicated EGFP-tagged DDX3X constructs for 24 h. After pulse labeling of newly synthesized proteins using puromycin for 30 min, cells were fixed and immmunostained against G3BP1 (magenta) and puromycin (PURO, red). EGFP signal was used to visualize the desired tagged proteins (denoted by white curve lines). Nuclei were stained with DAPI (blue). (d) Quantification of SGs in HeLa cells transfected with EGFP alone or the indicated EGFP-tagged DDX3X constructs for 24 h under normal conditions. Mean ± SEM values are based on a minimum of three replicated experiments. Student’s t-test comparison between full-length DDX3X and either ΔLCD1 or ΔLCD2 mutants is shown (***P ≤ 0.001). (e) Quantification of cells expressing EGFP plasmids (EGFP+) that are concomitantly devoid of PURO incorporation [PURO(−)] from the experiment shown in (c). Mean ± SEM values are based on a minimum of three biological replicates. Student’s t-test comparison between full-length DDX3X and either ΔLCD1 or ΔLCD2 mutants is shown (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant).