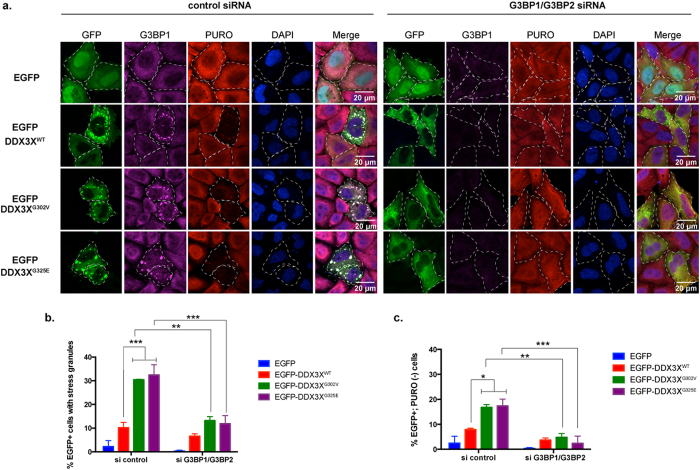
Figure 8. Knocking-down the SG nucleator factors G3BP1/G3BP2 prevents DDX3X-induced SG formation and allows normal rates of mRNA translation.
(a) Puromycin incorporation assay in HeLa cells treated with control siRNAs (left panel) or G3BP1 and G3BP2 siRNAs (right panel) for 24 h followed by transfection with the indicated EGFP plasmids for another 24 h. Immunofluorescence was performed against G3BP1 (magenta) and puromycin (PURO, red). GFP signal was used to visualize the desired tagged proteins. Shown are representative images for each sample. Dashed lines denote cells displaying DDX3X granules and are devoid of PURO incorporation. Nuclei were stained with DAPI (blue). (b) Quantification of SG formation in cells expressing EGFP (control) or EGFP-DDX3X variants (wild-type, G302V, or G325E) from the experiment shown in (a). Error bars represent mean values ± SEM (N = 3, Two-way ANOVA; **P ≤ 0.01, ***P ≤ 0.001). (c) Quantification of cells expressing EGFP plasmids (GFP+) that are concomitantly devoid of PURO incorporation [PURO(−)] from the experiment shown in (a). Error bars represent mean values ± SEM (N = 3, Two-way ANOVA; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).