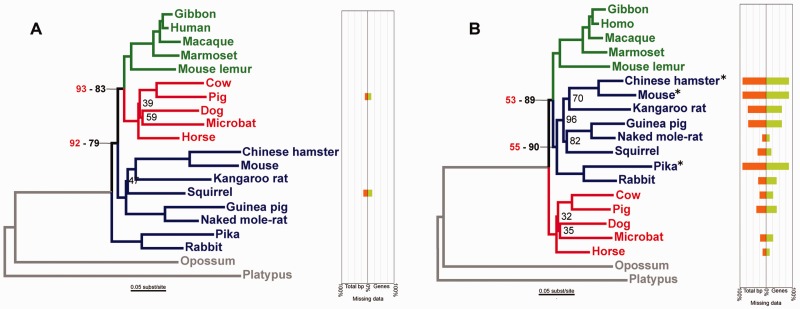
Fig. 3.
ML phylogenies and missing data plots obtained with (A) the unprocessed OL + 2 dataset, and (B) the OLLS³+2 dataset, for which data flagged by LS³ has been removed. Bootstrap support values <100 are presented in black, and support values for the same node in the ML trees obtained with the OL and OLLS³ dataset, which had the same topological arrangement of the three lineages of interest, are presented in red. Primates are colored green, Laurasiatherians are colored red, and Glires are colored dark blue. When using the unprocessed OL + 2 dataset (A), the Euarchontoglires group (Primates and Glires) is not recovered as monophyletic and Glires is recovered as paraphyletic. Upon removing the LS³-flagged sequences and genes from the dataset, the correct topology is recovered (B). Note that in (B), the bootstrap support from the OLLS³ dataset (in red) is much lower compared with that of the OLLS³+2 dataset (corresponding number in black). The missing data plots show the percent of total missing data by species (orange) and the percent of genes in which the species was not represented in the dataset (green). Asterisks (*) mark species that were eliminated from the OLLS³ dataset because they disrupted the lineage rate homogeneity in all genes. The scientific names of the species are as follows: Gibbon = Nomascus leucogenys; Human = Homo sapiens; Macaque = Macaca mulatta; Marmoset = Callithrix jacchus; Mouse lemur = Microcebus murinus; Cow = Bos taurus; Pig = Sus scrofa; Dog = Canis lupus familiaris; Microbat = Myotis lucifugus; Horse = Equus caballus; Chinese hamster = Cricetulus griseus; Mouse = Mus musculus; Kangaroo rat = Dipodomys ordii; Squirrel = Ictidomys tridecemlineatus; Guinea pig = Cavia porcellus; Naked mole-rat = Heterocephalus glaber; Pika = Ochotona princeps; Rabbit = Oryctolagus cuniculus; Opossum = Monodelphis domestica; and Platypus = Ornithorhynchus anatinus.