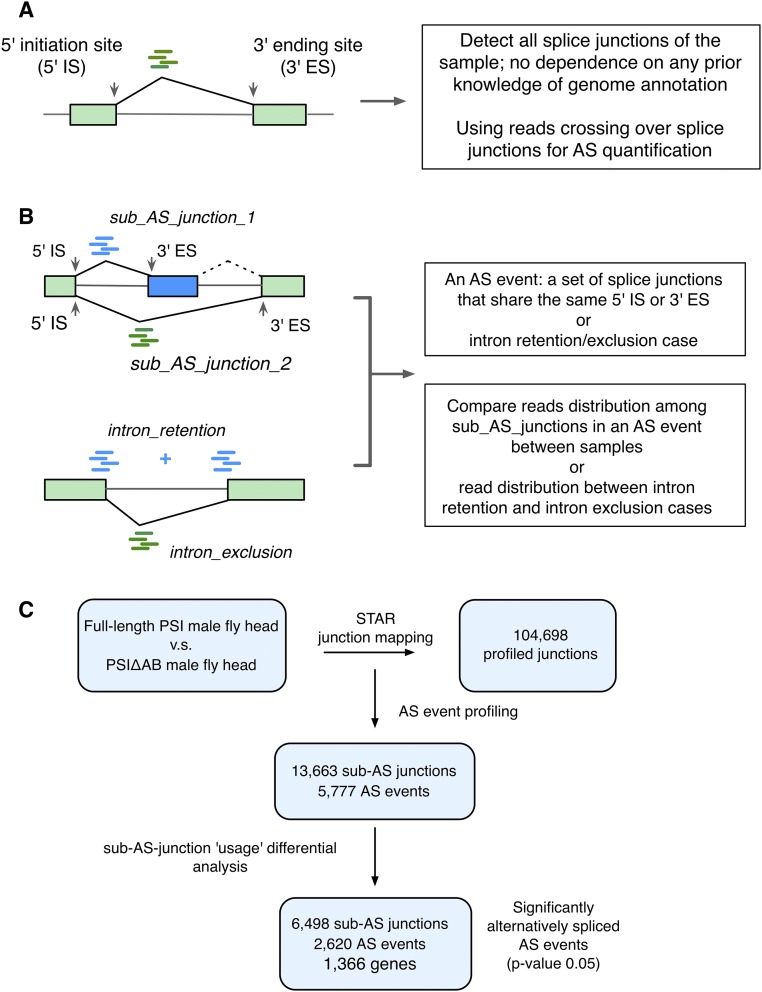
Fig. S2.
The computational pipeline for analyzing differential AS profiles using the JUM method. See Materials and Methods for details. (A) The JUM uses reads that cross over splice junctions for AS detection and quantification. (B) The JUM defines a nonintron-retention AS event as a set of splice junctions that share the same 5′IS or 3′ES. The use of each subjunction (read distribution to each subjunction) in the AS event is then compared among different experimental samples. For intron retention, read distribution between the retention and intron-exclusion cases are compared among different biological samples. (C) Flowchart of the JUM computational analysis to identify significantly changed AS targets of PSIΔAB from RNA-seq samples of full-length PSI and PSIΔAB mutant fly heads.