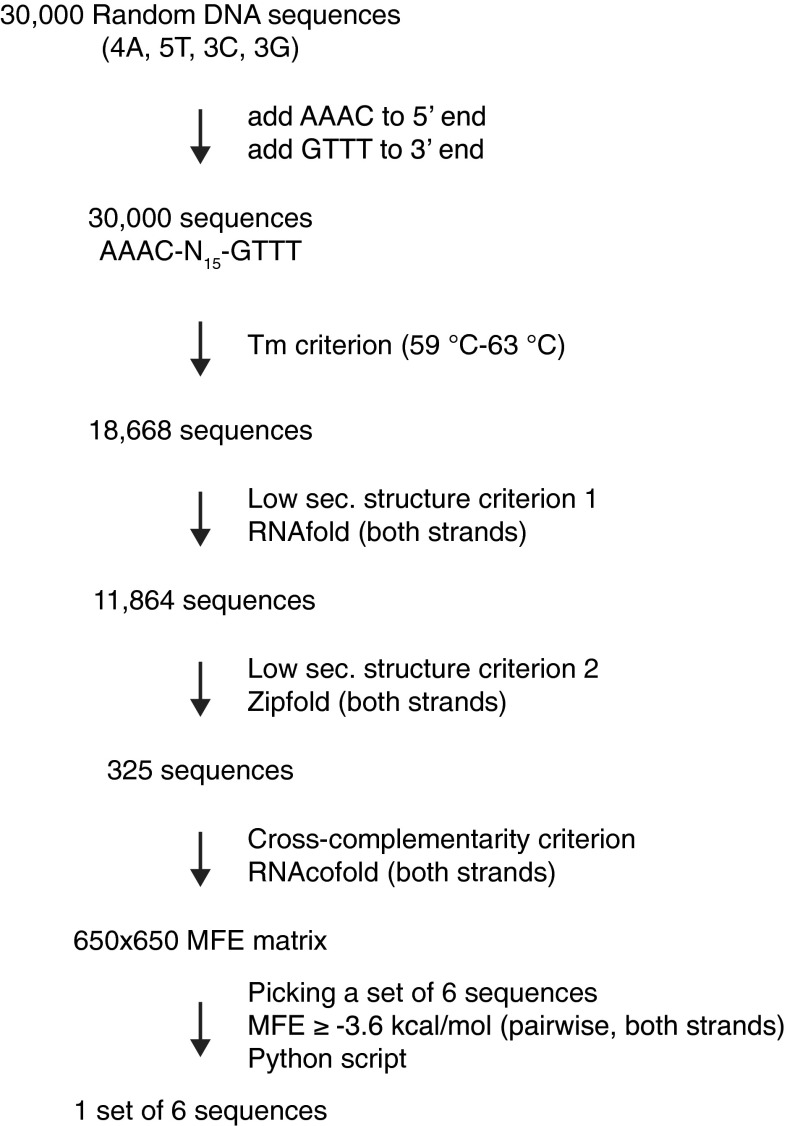
Fig. S1.
Generation of optimized DNA linker sequences. To generate optimized homology sequences for a six-fragment Gibson assembly reaction, 30,000 random DNA sequences of 15 nt in length and defined base composition were generated, and the sequences AAAC and GTTT required for compatibility with PmeI digestion were added to their 5′ and 3′ ends, respectively. Melting temperatures were predicted, and only sequences within the melting temperature range of 59–63 °C were included in further analyses (18,668 sequences). Sequences with a high probability of secondary structure formation on either strand were excluded using RNAfold (11,864 sequences left). The sequence set was further analyzed for secondary structure formation with Zipfold using parameters that fit with Gibson reaction conditions. Only sequences for which both strands were very unlikely to form secondary structures in Gibson assembly reactions [minimum free energy (MFE) ≥ +1.5 kcal/mol] remained in the sequence set (325 sequences). To pick a set of sequences that provides highest specificity in Gibson reactions (i.e., no false annealing), hybrid structures for each combination of two sequences out of the set of 325 sequences and their reverse complements were predicted using RNAcofold. Free energy values of the MFE structure of each hybrid were entered into a 650 × 650 MFE matrix. An algorithm was written in python to build sequence sets in which no pairwise interactions violate an MFE threshold. When setting the threshold to MFE ≥ –3.6 kcal/mol, it was possible to pick exactly one set of six sequences. This set is used as linker sequences in the second assembly step (linkers A, B, C, D, E, and F). This set was manually slightly modified to generate a set that is applicable in the first assembly step (linearization with SwaI), while maintaining all thermodynamic criteria (linkers α, β, γ, δ, ε, and ω).