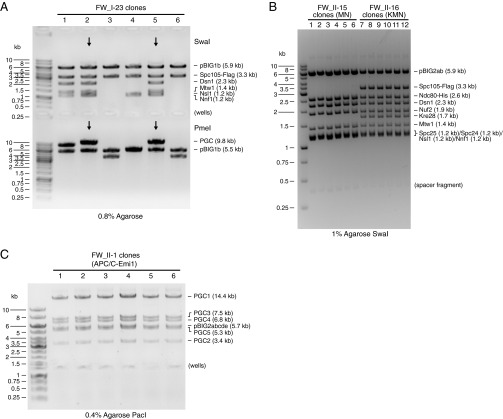
Fig. S5.
Examples for construct analysis. (A) Analytical digest of a pBIG1 construct. Six clones of FW_I-23 (pBIG1b:Dsn1/Mtw1/Nnf1/Nsl1/Spc105-Flag) were digested by SwaI or PmeI and analyzed on a 0.8% agarose gel. The expected sizes of pBIG1 vector backbone and GECs after SwaI digestion and of pBIG1 vector backbone and the PGC after PmeI digestion are indicated. Note that spacer sequences (see Fig. S3) stay with the vector backbone after SwaI digestion (5.9 kb) and with the PGC after PmeI digestion (backbone 5.5 kb). Clones 2 and 5 show the correct restriction pattern. (B) Analytical SwaI digest of pBIG2 constructs. Six clones of FW_II-15 (Kinetochore MN) and six clones of FW_II-16 (Kinetochore KMN) were digested with SwaI and analyzed on a 1% agarose gel. The expected sizes of pBIG2 vector backbone and GECs are indicated. Note that SwaI digestion of pBIG2 constructs yields an additional fragment of 0.3 kb consisting mainly of spacer sequences between PGCs. All clones show the correct restriction pattern. (C) Analytical PacI digest of a pBIG2 construct. Six clones of FW_II-1 (APC/C-Emi1) were digested with PacI and analyzed on a 0.4% agarose gel. The expected sizes of pBIG2 vector backbone and PGCs are indicated. All clones show the correct restriction pattern.