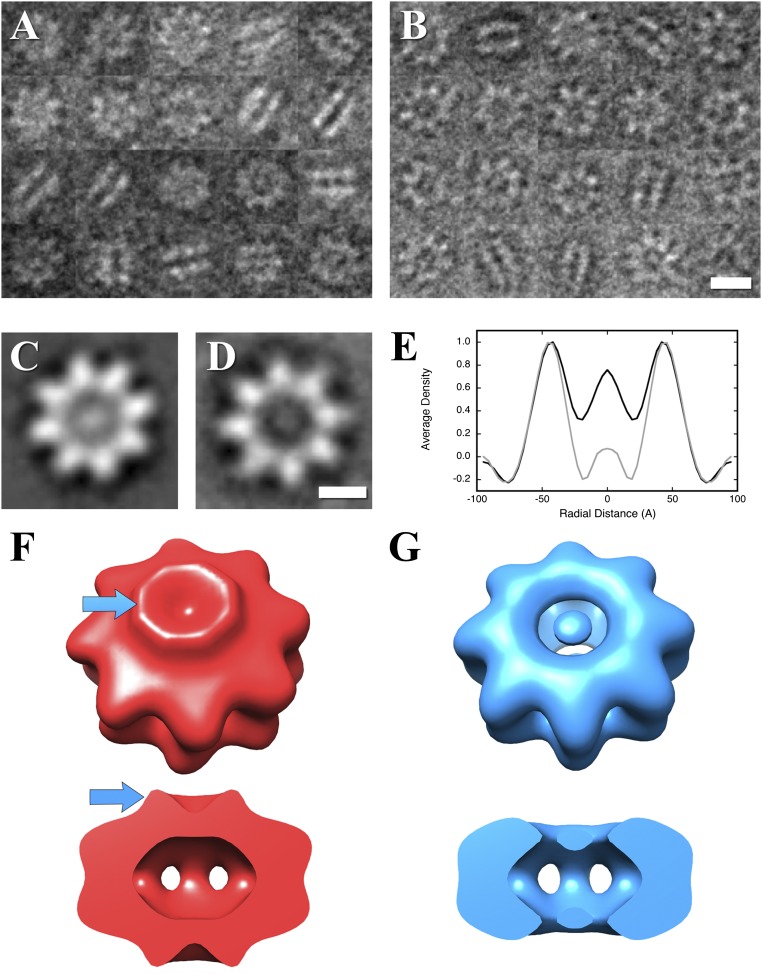
Fig. S3.
The RS1 domain does not affect oligomerization. (A) Negative-stain electron micrograph images of RS1 wild-type. (B) Negative-stain electron micrograph images of an N-terminal deletion mutant, RS1-∆N. (Scale bar, 100 Å.) (C) Two-dimensional class average of wild-type top views, showing a central density. (D) Two-dimensional class average of mutant top views without a central density. (Scale bar, 50 Å.) (E) Radial average profiles of the wild-type and mutant top views show that the density in the center of the mutant protein (radius 0–20 Å) is similar to the background (radius 70–95 Å). Side views in A and B show both wild-type and mutant to be double-ring structures for the great majority of the particles. (F) Reconstruction of the wild-type particle showing the RS1 domains as a ridge (blue arrows). (G) Reconstruction of the N-terminal deletion mutant, RS1-∆N, showing the absence of the RS1 domains.