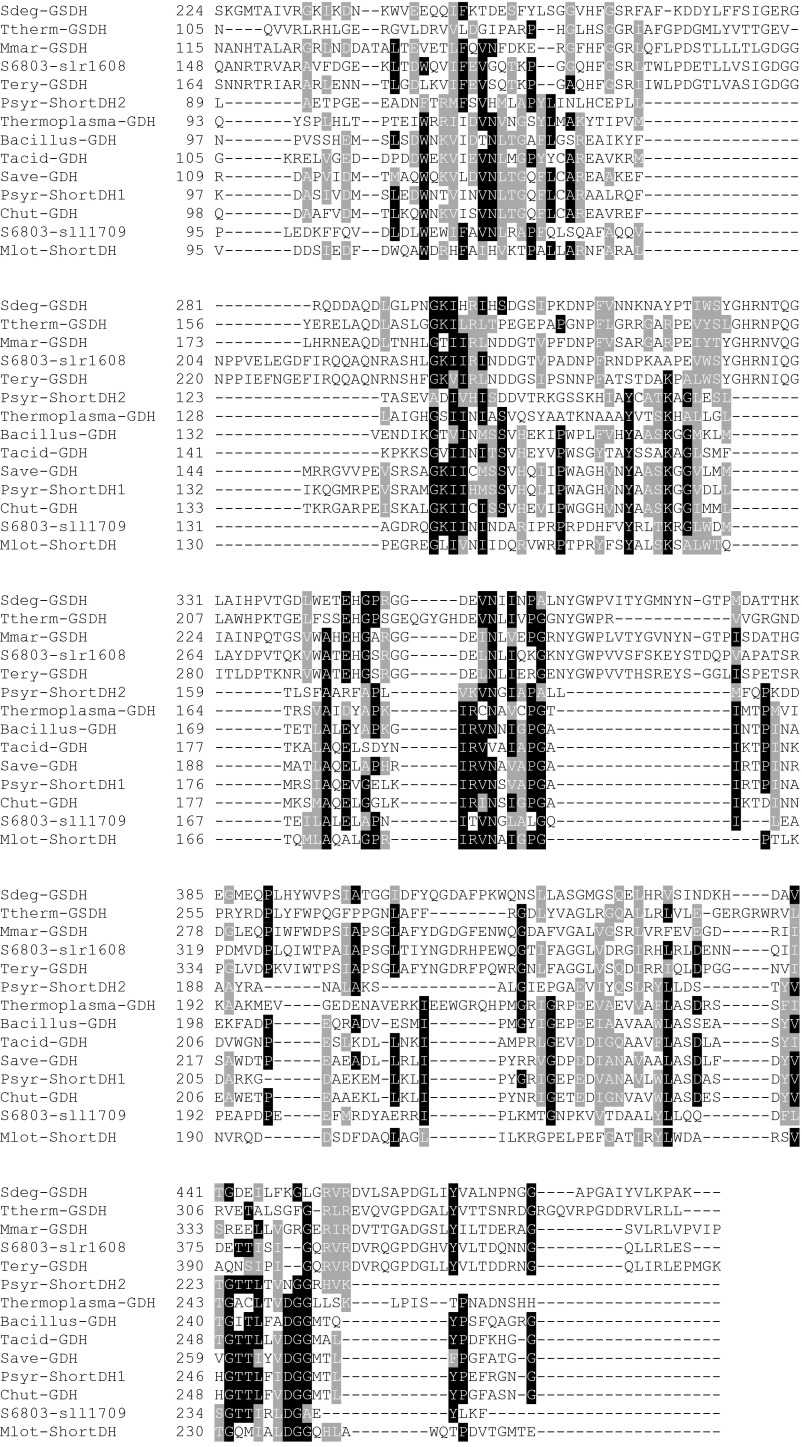
Fig. S3.
Alignment of Gdh, short-chain dehydrogenases, and glucose/sorbosone dehydrogenases with the two putative candidate proteins of Synechocystis sp. PCC 6803. The different proteins are from Saccharophagus degradans (Sdeg-GSDH), Thermus theromophilus Hb8 (Ttherm-GSDH), Maricaulis maris MCS10 (Mmar-GSDH), Trichodesmium erythraeum IMS101 (Tery-GSDH), Pseudomonas syringae pv. phaseolicola 1448A (Psyr-ShortDH2), Thermoplasma volcanium (Thermoplasma), Bacillus megaterium (Bacillus-GDH), Thermoplasma acidophilum (Tacid-GDH), Streptomyces avermitilis MA-4680 (Save-GDH), Pseudomonas syringae pv. syringae B728a (Psyr-ShortDH1), and Mesorhizobium loti MAFF303099 (Mlot-ShortDH). Structural information can be obtained from the Protein Data Bank (www.rcsb.org/) for the proteins of B. megaterium, Th. acidophilum, and T. thermophiles Hb8 under the IDs 1GCO, 3VTZ, and 2ISM, respectively. The genome of Synechocystis was searched for a Gdh. Two candidate genes could be identified. Sll1709 is annotated in the data bank cyanobase as Gdh and/or alternatively as 3-ketoacyl-acyl carrier protein reductase. It contains the Rossman fold typical for dehydrogenases binding NAD(P)+. The highest similarities are to the short-chain dehydrogenases (3-ketoacyl-acyl carrier protein reductase) of P. syringae and the Gdhs of a number of prokaryotes. The second candidate is the gene slr1608, which is annotated as a putative periplasmic Gdh in cyanobase. The closest homologs of this gene in other cyanobacteria are annotated as glucose/sorbosone dehydrogenases. These proteins do not contain a Rossmann-fold, so they do not use NAD(P)+ as substrate. According to the Signa lP tool of the Center for Biological Sequence Analysis prediction server (www.cbs.dtu.dk/services/), it is localized to the periplasm with a cleavage site between position 20 and 21 between the two Serin residues in the sequence ALS-SCG. Its localization indicates that it might reduce plastoquinone when oxidizing glucose. Deletion of sll1709 in ΔzwfΔpfk inhibited the ability of the mutant to enhance its growth on mixotrophic conditions (Fig. 2B).