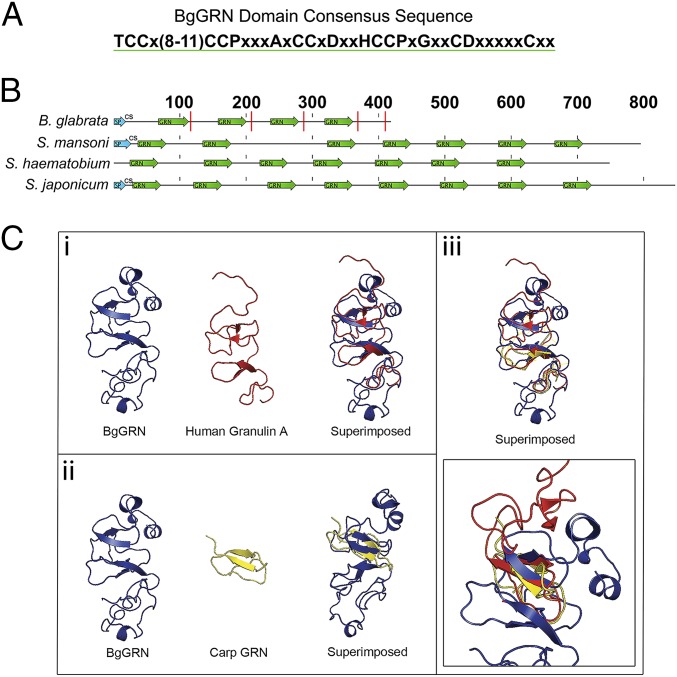
Fig. 1.
In silico prediction of BgGRN granulin domain architecture compared with some known and predicted granulins. (A) The consensus sequence of the BgGRN granulin domain highlighting the canonical 12-cysteine motif that defines granulin proteins. (B) Reference diagram highlighting the predicted locations of the granulin domain (green arrows) in the predicted progranulin protein sequences of B. glabrata, S. mansoni, S. hematobium, and S. japonicum. Predicted signal peptides (SP) are highlighted in blue arrows and are followed by the predicted cut site (CS) for the signal peptide. Predicted elastase cleavage sites for BgGRN are highlighted with a red line intersecting the protein at the approximate cut site. (C) The predicted 3D structure of BgGRN compared with the two known crystal structures for human (i) and carp (ii) granulins, with a focus on the similar organization of β-sheets and α-helices (iii).