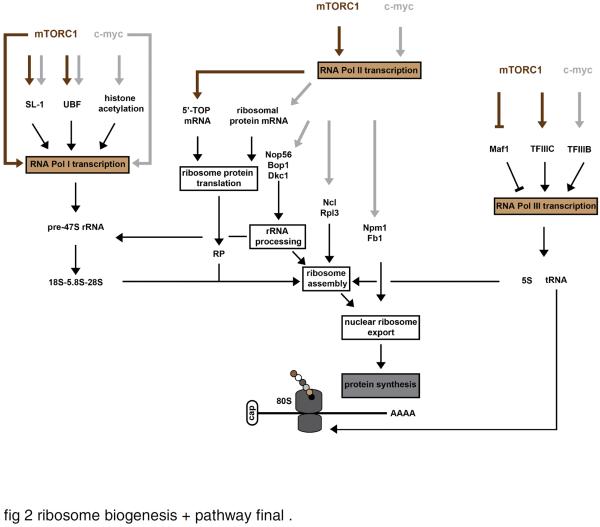
Fig. 2.
Regulation of ribosome biogenesis by mTORC1 and c-myc.
mTORC1 (mechanical target of rapamycin complex 1) and c-myc (c-myelocytomatosis oncogene) work in concert with each of the three RNA polymerases in regulating ribosome biogenesis. mTORC1 and c-myc seem to directly promote the transcription of the pre-rRNA 47S, and can lead to the activation of the RNA Pol I transcriptional factor SL-1 (selectively factor-1), and the rDNA transcription factor UBF (upstream binding factor). Some lines of evidence also suggest that c-myc indirectly increases the RNA Pol I transcription by promoting the opening of the chromatin structure near rDNA loci through histone acetylation. mTORC1 appears to regulate the translation of 5'-TOP (terminal tract of pyrimidine) mRNAs which encode ribosomal proteins and other components of the translational machinery, while c-myc activates the transcription of several ribosomal protein-encoding genes. In addition, c-myc promotes the transcription of several auxiliary factors required for ribosome biogenesis, such as genes involved in rRNA processing [Nop56 (Nop56 ribonucleoprotein), Bop1 (block of proliferation 1) and Dkc1 (dyskeratosis congenital 1, dyskerin)], ribosome assembly [Ncl (nucleolin) and Rpl3 (ribosome protein L3)], and nuclear ribosome export [Npm1 (nucleophosmin 1) and Fbl (fibrillarin)]. Finally, mTORC1 and c-myc activate the RNA Pol III transcription through their interaction with TFIIIC (transcription factor IIIC) and TFIIIB (transcription factor IIIB), respectively. mTORC1 may also inhibit the function of the RNA Pol III repressor Maf1.