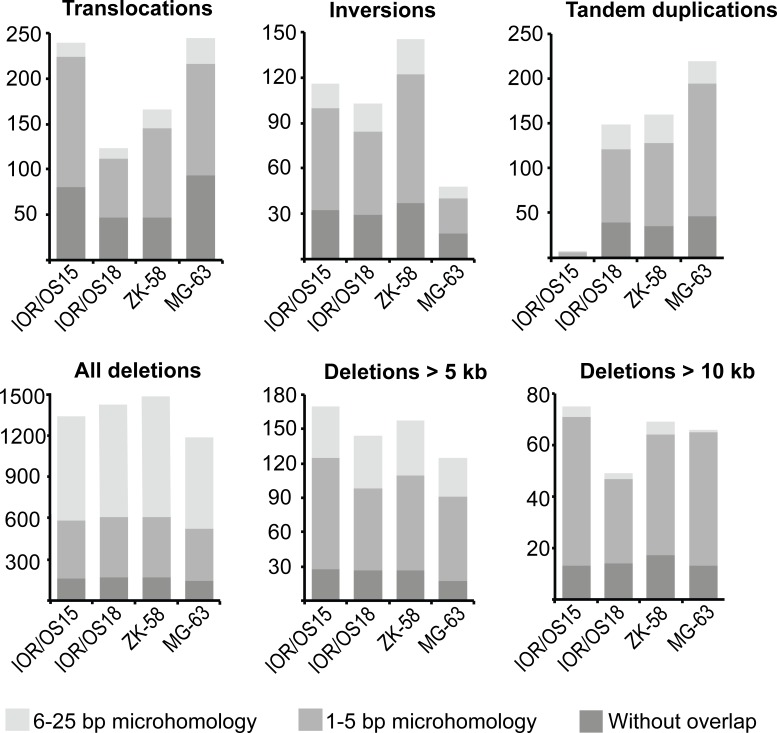
Figure 2. Abundance of the different genomic rearrangement types and their microhomology pattern.
The rearrangements are grouped by type (translocations, inversions, tandem duplications, deletions) and divided in three categories of overlapping microhomology at the breakpoints: 6-25 bp overlap, 1-5 bp overlap and without overlap. Deletions are further divided by length, showing that smaller deletions (< 5 kb) have a different microhomology pattern with a higher frequency of 6-25 bp long overlapping microhomology. This frequency was clearly reduced for deletions longer than 5 kb and more similar to the other types of rearrangements. The same tendency was found for deletions longer than 10 kb, which showed even more similarity to the microhomology pattern of translocations, inversions and tandem duplications.