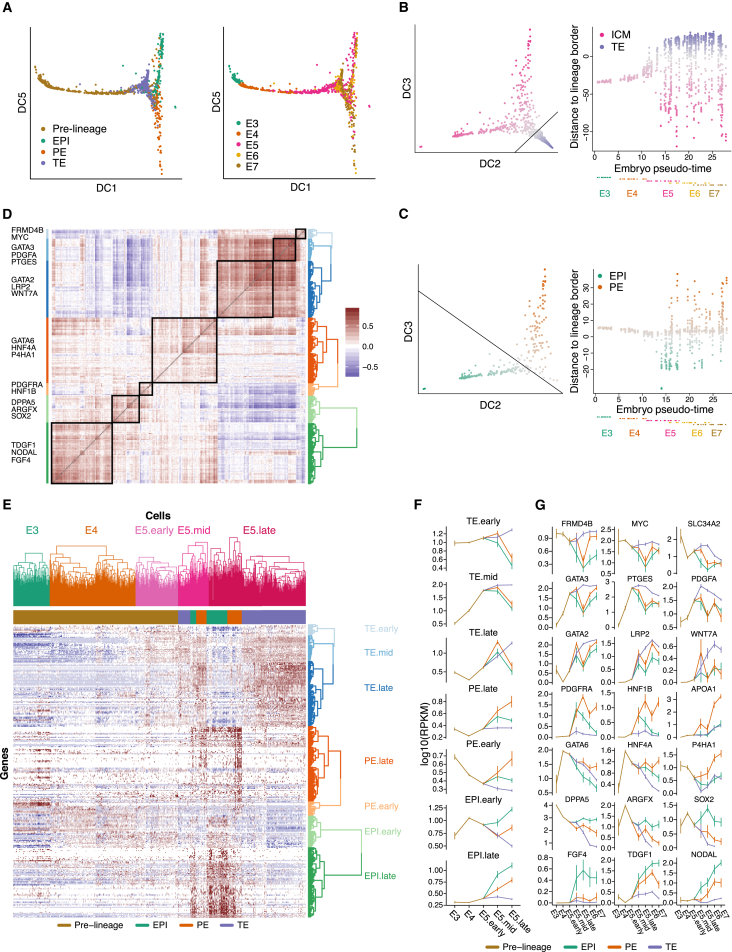
Figure 4.
Developmental Progression from E3 to E7 Showing the Formation of Blastocyst Lineages
(A) Three-dimensional diffusion map representation of all cells, showing lineage assignment and embryonic day, respectively. A total of 94 lineage-specific genes at E5 were used as input (Supplemental Experimental Procedures). DC, diffusion component.
(B) Lineage segregation of all 1,529 cells with respect to ICM versus TE. Left: the expression of every cell with respect to lineage-specific genes (axis represent diffusion-components [DC], analogous to principal components). The black line depicts a lineage-separating border that optimally separates the two classes of cells, determined by a support vector machine (Supplemental Experimental Procedures). Right: the y axis indicates the distance from the lineage decision boundary (black line in the left sub-figure). The x axis indicates pseudo-time, as determined in Figure 1F. Each embryo was assigned a time using the mean of the cellular pseudo-times of the cells in that embryo. Each dot below the x axis indicates an embryo, colored by the embryonic day of sampling.
(C) As (B) but with respect to EPI versus PE.
(D) Gene-gene Pearson’s correlation matrix using the top 100 lineage-specific genes from each lineage. Gene-modules were determined based on hierarchical clustering of the correlation matrix and labeled with representative genes being part of the cluster.
(E) Heatmap of expression levels (RPKM) for E3–E5 cells using the top 100 lineage-specific genes from each lineage. Cell groups were ordered according to their pre-determined groups, indicated by the colored dendrogram, and clustered within their respective group (E3, E4, E5.early, E5.mid, and E5.late). E5.mid cells were classified into three sub-groups based on the observed hierarchical clusters (EPI, PE, and TE). Genes were grouped according to observed hierarchical clusters and named based on which type of cells, and at which time point, the genes were expressed.
(F) RPKM mean expression levels of lineage-specific gene sub-clusters as identified in Figure 4D. Vertical lines indicate 95% non-parametric bootstrap confidence interval across cells (B = 1,000).
(G) RPKM expression levels of representative genes from each gene sub-cluster. Vertical lines indicate 95% non-parametric bootstrap confidence interval across cells (B = 1,000).