Figure S2.
Identification of the Most Variable Genes and Temporal Separation of Preimplantation Single-Cell Transcriptomes, Related to Figure 1
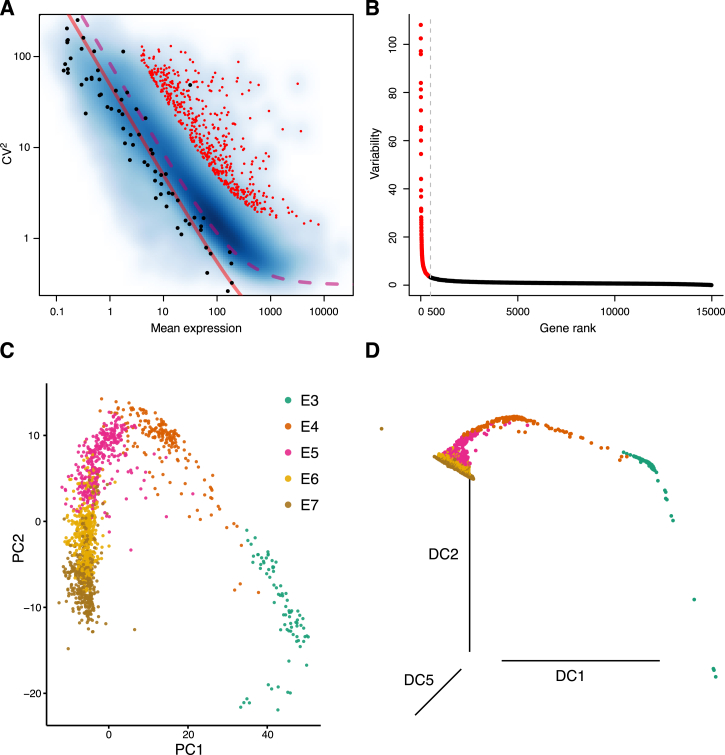
(A) Gene mean expression versus squared coefficient of variation (CV2) of all RefSeq genes. Red dots indicate genes ranked as being among the 500 most variable genes. Black dots indicate ERCC spike-in transcripts. Red line represents a fit against the ERCC transcripts, indicating the technical variability, and dotted line represents a biological variability of CV = 0.5, added to the technical one.
(B) Variability test-statistic of every expressed RefSeq gene, derived from the mean-variance relationship in Figure S1A (Supplemental Experimental Procedures), versus the gene-rank; where rank was obtained by ordering by the variability test-statistic. Red dots indicate genes ranked as being among the 500 most variable genes.
(C) Principal component analysis of all 1,529 cells using the 500 most variable genes.
(D) Diffusion map of all 1,529 cells using the 500 most variable genes.