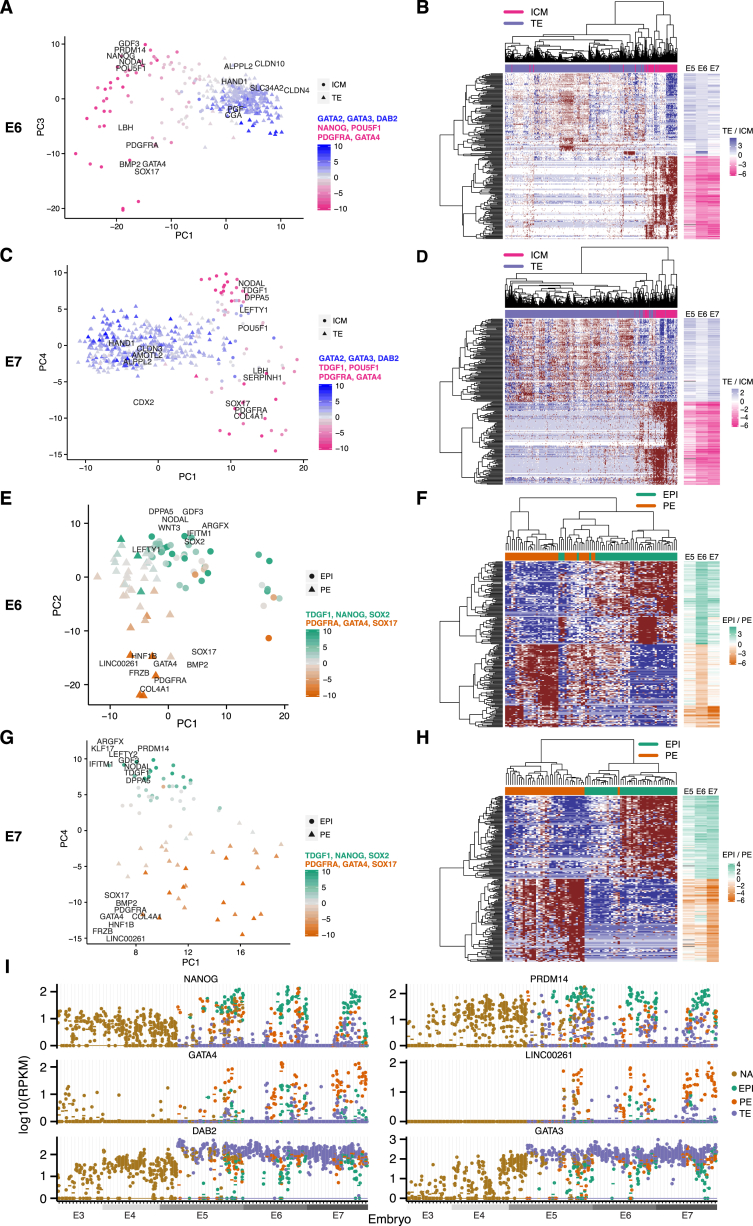
Figure S3.
Lineage Segregation of Cells into Inner Cell Mass, Trophectoderm, Epiblast, and Primitive Endoderm, Related to Figure 2
(A) PCA biplot showing ICM and TE classification of cells from E6. Cells were classified as ICM or TE using PAM clustering in the PCA dimensionality-reduced space with the 250 most variable genes across all E6 cells as input (Supplemental Experimental Procedures). Genes with high PC loadings are shown. Colors indicate the weighted mean of the expression of known lineage markers, listed above the color bar, using weights −1 and 1 for ICM and TE genes, respectively.
(B) Heatmap of E6 cells and the top 200 differentially expressed genes between ICM and TE E6 cells (top 100 genes from each lineage). The upper colored bar indicates lineage-classification of each cell, as determined in (A). Right-hand-side bars indicate the log2 fold-change of the TE divided with ICM mean-expression level for each gene and embryonic day (E5-E7).
(C and D) As in (A) and (B) but with respect to E7 cells.
(E and F) As in (A) and (B) but with respect to E6 ICM cells, contrasting EPI and PE cells.
(G and H) As in (E) and (F) but with respect to E7 ICM cells, contrasting EPI and PE cells.
(I) Expression for a selection of top-ranked lineage-specific marker genes stratified by embryo and lineage. Each dot represents the expression in a single cell and vertical lines segregate embryos. Horizontal lines indicate mean expression level per lineage within each embryo.