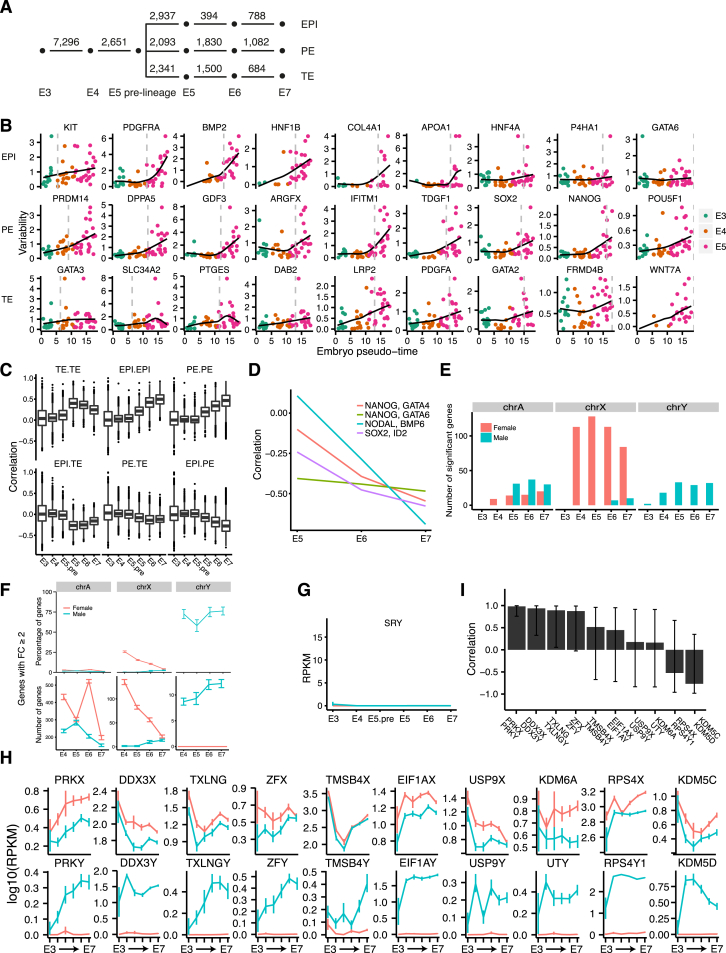
Figure S4.
Preimplantation Developmental Progression of Lineage-Specific and Sex-Specific Genes, Related to Figure 4
(A) The number of significantly differentially expressed genes between embryonic time-points. From E5 to E7 the differential expression analysis was done within lineages.
(B) Gene expression variability within each embryo versus developmental time (Supplemental Experimental Procedures). Each dot represents an embryo.
(C) Gene-gene Pearson correlations among the top 300 maintained lineage genes (100 from each lineage). Titles refer to that genes specifically expressed in each of the two listed lineages were correlated to each other.
(D) Pearson correlation within ICM cells against developmental stage for gene-pairs selected among lineage-specific genes with the strongest anti-correlation.
(E) The number of significantly differentially expressed genes between females and males at each embryonic day, stratified by the genes’ chromosomal location: autosome (chrA), chromosome X (chrX) and chromosome Y (chrY)
(F) The number and percentage of genes with fold-change (FC; female versus male cells) ≥ 2. Error-bars indicate standard deviation obtained by bootstrap resampling of cells (n = 100). Red and blue lines represent genes with higher expression in female and male, respectively.
(G) RPKM expression levels of the testis-determining factor SRY.
(H) RPKM stage-wise mean expression levels of X- and Y-linked paralogous gene pairs that were significantly differentially expressed. Error-bars indicate 95% confidence interval.
(I) Pearson correlation between male stage-wise mean expression levels of X- and Y-linked paralogous gene pairs. Error-bars indicate 95% confidence interval.