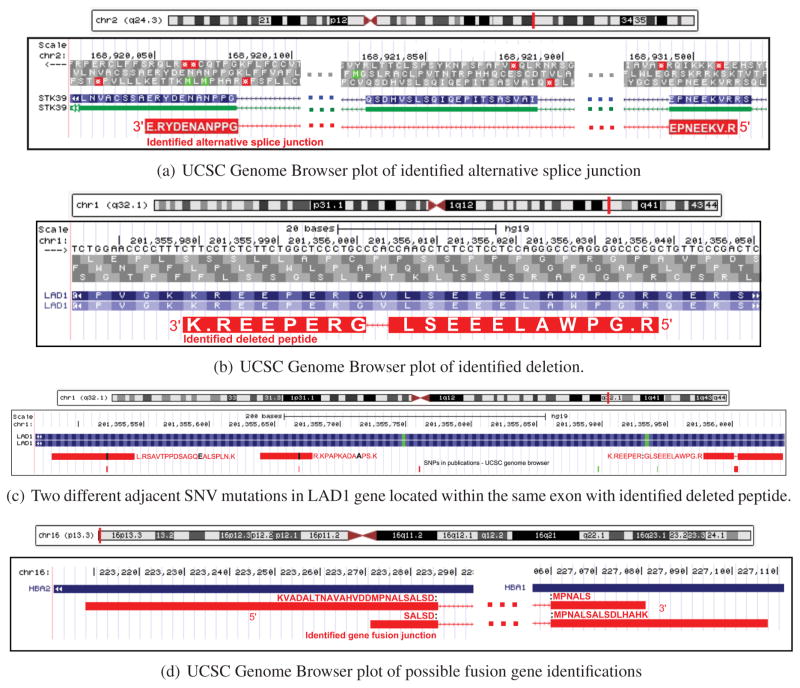
Figure 6.
(a) Identified alternative splice junction peptide. Peptide VKEENPE:G:PPNANEDYR (junction existing in the middle of amino acid G) had 11 spectra counts (with unique genomic location), and a total 386 RNA-seq reads were mapped to this alternative splice junction. (b) Identified deletion and two neighboring SNP mutated peptides. This peptide with the deletion had 7 spectra counts (across 6 different tumor protein samples) with unique genomic location and 996 RNA-seq read depth (across 10 different tumor DNA samples). (c) Two additional SNV mutations within the exon of the above deletion example. All mutations had external supporting evidence from dbSNP. SNV mutation of the peptide K.NLPSLA:E:QGASDPPTVASR.L (K → E) was also reported by TCGA3 colon cancer somatic mutation calls, with a read depth of 10 711. (d) Identified fusion gene peptides. This shows a possible gene fusion region where two junctional peptides are identified accross two different genes (HBA1 and HBA2). Two fusion peptides shown in this region had unique genomic locations and a total of 15 spectra counts. HBA1 and HBA2 are hemoglobin-related genes.