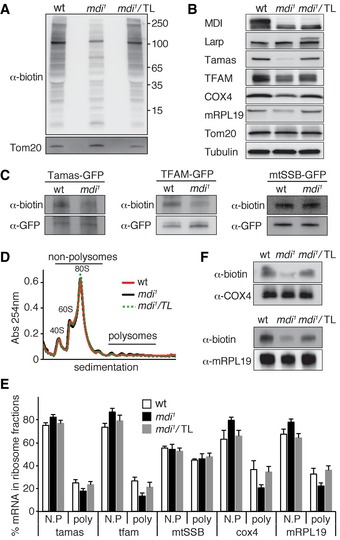
Detection of nascent protein synthesis in the mitochondrial fraction of the ovary. Nascent protein synthesis was monitored by AHA incorporation and detected by anti‐biotin antibody. Tom20 served as a loading control. Note that the synthesis of nuclear‐encoded mitochondrial proteins was decreased in the mdi
1 background and was restored by overexpressing Tom20‐Larp (mdi
1/TL).
Western blots of mitochondrial proteins in ovarioles of wt, mdi
1, and mdi
1 flies expressing Tom20‐Larp (mdi
1/TL) fusion protein. Note that levels of Tamas, TFAM, mRpL19, and COX4 were reduced in mdi
1 flies, but restored in mdi
1/TL flies.
Nascent protein synthesis of Tamas and TFAM was decreased in mdi
1 ovary, whereas mtSSB was not affected. Tfamgfp, Tamasgfp, and mtSSBgfp were expressed in wt or mdi
1 background. Nascent proteins were labeled by AHA incorporation, and then, the GFP‐tagged protein was immunopurified with a GFP antibody and the nascent protein synthesis was detected by anti‐biotin antibody.
Representative profile of 254 nm absorbance of wt, mdi
1, and mdi
1/TL ovary extracts.
Polysome mRNA profiling for tamas, tfam, mtSSB, cox4, and mRPL19 in wt, mdi
1, and mdi
1/TL ovary. The percentage of mRNA for each gene in non‐polysomal fractions (N.P, including ribosomal subunits and monosome‐associated) and polysomal fractions (poly) was calculated and plotted. The fractions of tamas, tfam, cox4, and mRPL19 mRNAs in the polysomal fractions were significantly decreased in mdi
1 compared to wt, but were restored in mdi
1/TL flies. N = 4 for all samples. P‐values of comparing wt to mdi
1: tamas, P = 0.0055; tfam, P = 0.001; cox4, P = 0.0066; mRPL19, P = 0.0097. P‐values of comparing mdi
1/TL to mdi
1: tamas, P = 0.0206; tfam, P = 0.0346; cox4, P = 0.0036; mRPL19, P = 0.0016.
Nascent protein synthesis of COX4 and mRPL19 was decreased in mdi
1 ovary, but restored by overexpressing Tom20‐Larp (mdi
1/TL). The proteins were immunopurified with antibodies against the endogenous proteins.