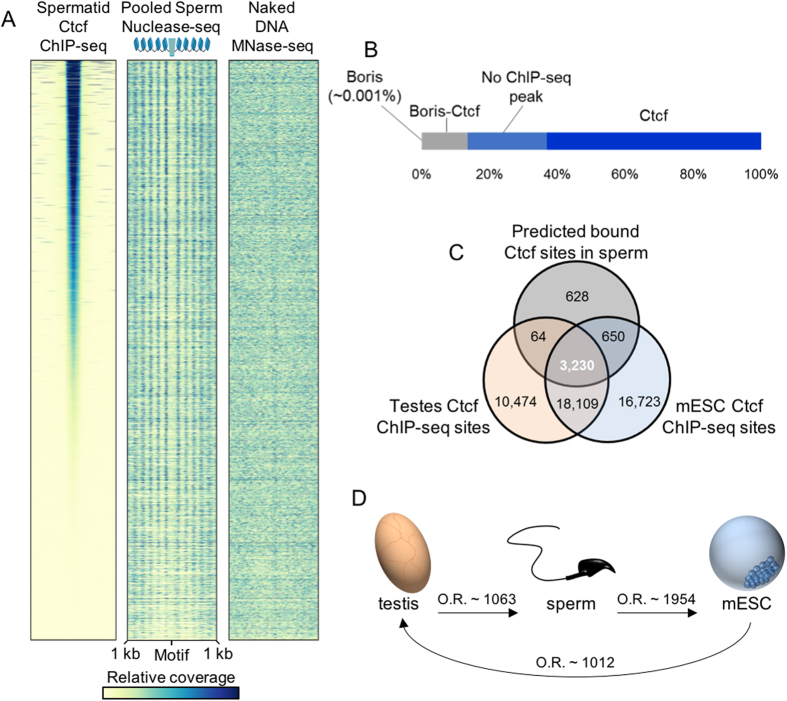
Figure 8. Ctcf remains bound in mature sperm following chromatin remodeling in mouse.
(A) Heatmaps of relative sequencing coverage in 2 kb windows centered on the complete set of 7,967 Ctcf sites bound in mature mouse sperm (Methods). Data are presented for ChIP-seq against Ctcf in round spermatids22, sperm nuclease-seq, and digested naked DNA libraries (left, center, and right, respectively). Sites are ranked according to decreasing ChIP-seq coverage. (B) The majority of the Ctcf sites occupied in sperm (n = 7,967) are occupied solely by Ctcf in round spermatids (n = 5,034, 63.2%). A subset of sperm Ctcf sites correspond to co-occupied Boris-Ctcf peaks in round spermatids (n = 1,075, 13.5%; Supplemental Figure 8B) or lack a corresponding ChIP-seq peak in the prior cell type (n = 1,857, 23.3%). In the latter case this was due to peaks not achieving significance (Supplemental Figure 8A). (C,D) The majority of Ctcf sites (Motif M01200) bound in sperm are observed in mouse ENCODE testis and mESC ChIP-seq datasets. Odds ratios (O.R.) from Fisher exact tests are presented between cell types comparisons.