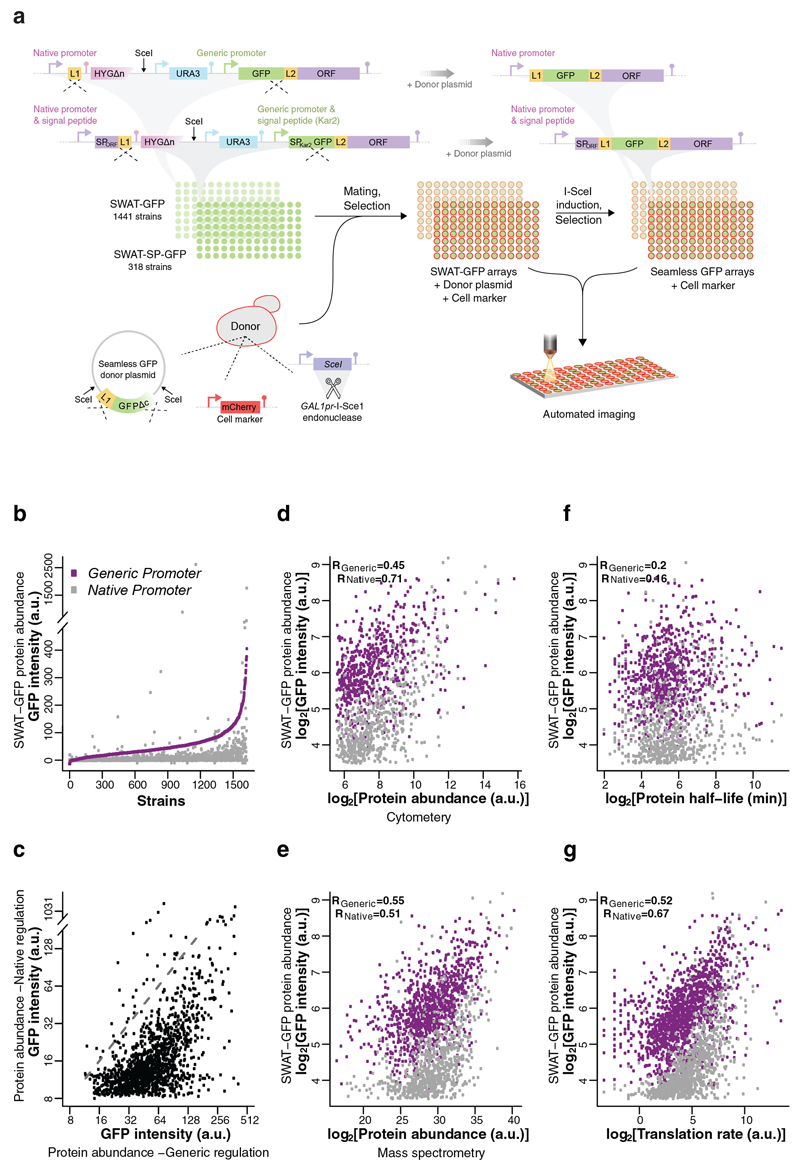
Figure 2. Utilizing SWAT strategy to rapidly create a seamless N′-tagged GFP library enables comparison of protein abundance under generic or native regulation.
(a)Two strain collections were assembled, in which proteins were tagged at their N′ with an acceptor SWAT module that contains the SpNOP1 constitutive promotor (generic) and a GFP. Proteins predicted to harbor a signal peptide (SP) were tagged with a similar acceptor SWAT module that contains a SP upstream to the GFP (SP of Kar2). The two libraries were crossed with a donor strain that harbors a plasmid with a seamless GFP swap donor plasmid, the inducible I-SceI enzyme and an mCherry cell marker. Imaging of strains was performed on both the parental SWAT-GFP library and the daughter seamless GFP library to compare protein abundance under generic or native regulation. (b) Expression levels of GFP-protein fusions under a single generic promoter spanned >2 orders of magnitude, and were almost exclusively higher than under native regulation. Purple: Generic regulation (SWAT-GFP); Grey: native regulation (seamless GFP). a.u - arbitrary units. (c) The correlation between protein abundance under generic and native regulation highlighted the effect of non-transcriptional regulation on protein abundance. Dashed line indicates the diagonal. (d-g) Comparison of N′ GFP tagged protein abundances, either under generic or native regulation with the: (d) abundance of these proteins as measured by flow cytometry analysis of C′ tagged GFP proteins27; (e) abundance of these proteins as measured by mass spectrometry 28; (f) protein half-lives 29; (g) protein translation rates as measured by ribosome profiling 30.