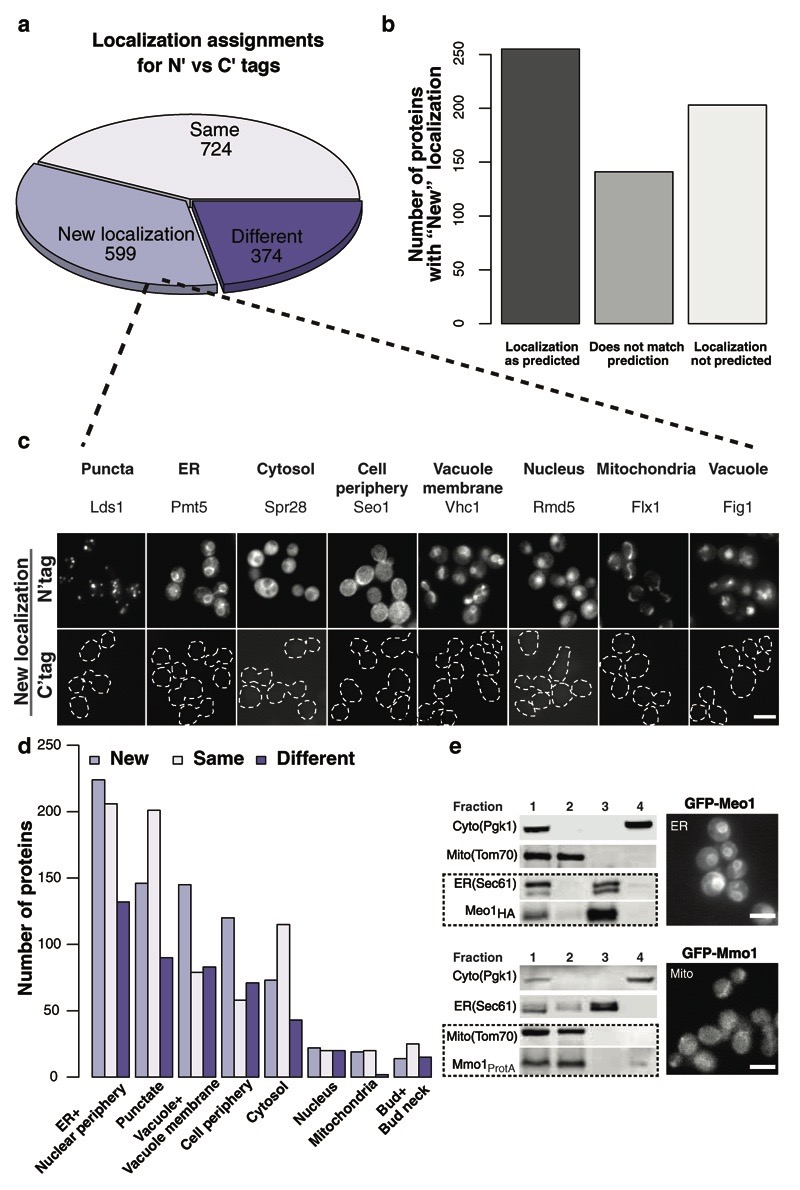
Figure 3. Creation of an N′ SWAT library sheds new light on hundreds of endomembrane proteins.
(a) Comparing protein N′ tagging localization assignments to assignments made from the C′ tagged library 4 showed that 724 proteins are visualized in the same localization (Same), 374 show different (Different) localization, and 599 proteins were only visualized using N′ tagging (New localization). For all localization assignments please see (Supplementary Table 4). (b) Comparing the 599 proteins that were only visualized using N′ tagging to GO ontology indicates that for nearly 30% no GO term was ever annotated. (c) A representative image for proteins that reside in each sub-cellular localization from the 599 proteins (New localization) that could be visualized by N′ tagging with the SWAT-GFP module (top) as opposed to C′ tagging with GFP (Bottom). Scale bars, 5 µm. (d) New assignments of protein localization by N′ tagging in the SWAT-GFP and SWAT-SP-GFP collections were mostly to the ER, punctate structures, the vacuole and cell periphery. Scale bars, 5 µm. (e) Two examples for the 78 small open reading frame proteins (smORFs) that were visualized for the first time using the SWAT-GFP libraries. Ybr126w-a was found to localize to the ER (top, newly named Meo1), and Ykl044w to mitochondria (bottom, named Mmo1). Their localization was validated by a cell fractionation assay in which Meo1 co-fractioned with the ER protein Sec61, and Mmo1 with the mitochondrial protein Tom70. Fraction numbers: 1-Post nuclear supernatant (PNS); 2-Pellet (P13); 3-Pellet (P100); 4-Supernatant (S100) (Online Methods).