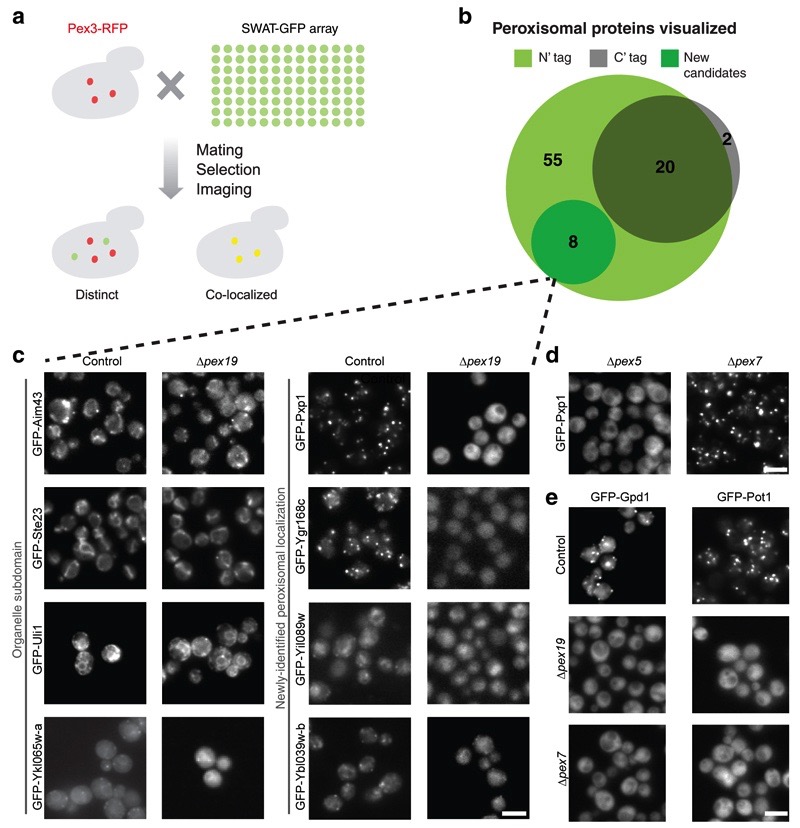
Figure 4. Correct targeting of peroxisomal proteins is maintained by the N′ GFP tagged strains enabling characterization of new peroxisomal proteins.
(a) The SWAT-GFP array was mated with a strain containing the peroxisomal marker Pex3-mCherry to query for peroxisome localized proteins. (b) Pex3 co-localization was observed in 55 proteins from the SWAT-GFP libraries, eight of which were never described before as peroxisomal (New candidates). Only 22 of these proteins were previously visualized by C′ GFP tagging, exemplifying the importance of an N′ GFP library. (c) Peroxisomal targeting of the eight new candidate proteins was validated by deletion of PEX19, which abolishes peroxisome biogenesis. Four proteins (left) remained in punctate formations, and are therefore most likely not targeted to peroxisomes, but rather to organelle sub-domains that reside near peroxisomes. Four other proteins (Right) dispersed to cytosolic localization upon PEX19 deletion validating them as bona fide peroxisomal proteins. Scale bars, 5 µm. (d) The prediction that Yel020c (now named Pxp1) is targeted to peroxisomes by a PTS1 was validated by deletion of the PTS1 receptor PEX5, in which Pxp1 no longer showed a peroxisomal localization (left), in contrast to deletion of the PTS2 receptor PEX7. Scale bars, 5 µm. (e) Protein targeting to peroxisomes via the N′ PTS2 signal was maintained despite the N′ SWAT-GFP tag. Gpd1 and Pot1 show proper localization to peroxisomes with the N′ SWAT-GFP tag (top). Their reliance on PTS2 for correct targeting is demonstrated by dispersion to the cytosol once the PTS2 receptor PEX7 is deleted (bottom), similar to the phenotype generated when peroxisomes are depleted completely by deletion of PEX19 (middle). Scale bars, 5 µm.