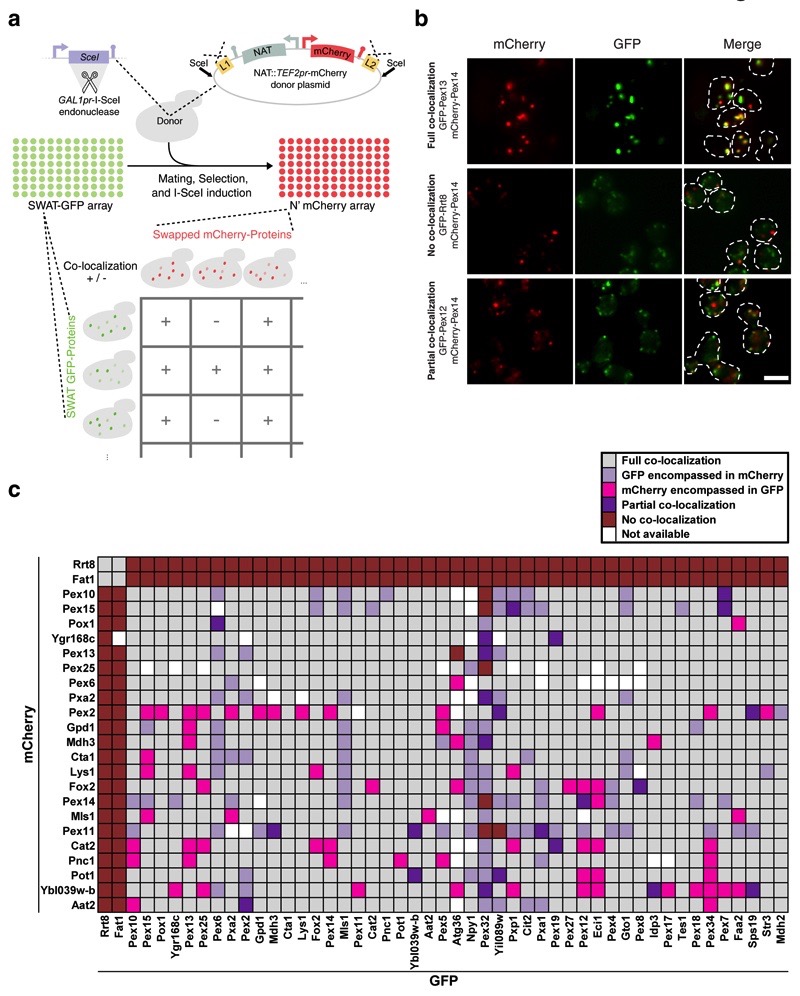
Figure 5. Rapid creation of a new N′ mCherry library exemplifies the swap-tag technology and opens up opportunities for systematic co-localization studies.
(a) An N′-mCherry tagged library was made by crossing the SWAT-GFP arrays with a donor strain containing a donor plasmid with the swap cassette NAT::TEF2pr-mCherry (Online Methods). Strains from the mCherry library, selected to be MAT alpha, can now be crossed with MATa SWAT-GFP strains to perform systematic co-localization experiments. (b) The co-localization experiment yielded several different patterns. Examples of three types of co-localization that were seen are given: a comprehensive overlap of the two proteins (top), distinct localization for each protein (middle), and a partial overlap (bottom). Scale bars, 5 µm. (c) Peroxisomal protein co-localizations were assayed in a systematic manner by mating of 24 mCherry tagged strains with 49 GFP tagged strains. While most peroxisomal proteins where visualized in the same peroxisomes (gray), in many instances proteins were visualized in separate groups of peroxisomes. The two lipid droplet proteins (Rrt8 & Fat1), which served as a negative control, indeed never co-localize with any of the peroxisomal proteins but did co-localize with each other.