Figure 2.
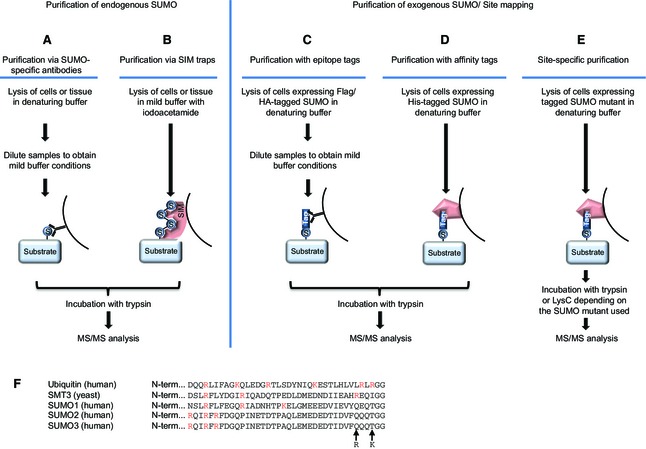
Proteomic approaches to identify SUMO targets. (A) Purification via SUMO‐specific antibodies. Cells are lysed under denaturing conditions to inactivate SUMO proteases. Afterwards, samples are diluted to obtain mild buffer conditions and SUMOylated proteins are purified using SUMO‐specific antibodies. Proteins are subsequently trypsinized and subjected to mass spectrometry. (B) Purification via SIM traps. Cells are lysed in a mild buffer supplemented with iodoacetamide, and SUMOylated proteins are purified using the SIM‐containing protein RNF4 as bait. Proteins are subsequently trypsinized and subjected to mass spectrometry. (C) Purification with epitope tags. Cells expressing a tagged SUMO fusion protein are lysed in denaturing buffer. For subsequent immunoprecipitation of the SUMO target proteins using antibodies targeting the protein tag, samples are diluted to obtain mild buffer conditions. Finally, they are trypsinized and analysed via mass spectrometry. (D) Purification with affinity tags. Cells expressing SUMO tagged with affinity tags are lysed in denaturing buffer, and SUMO targets are purified using affinity matrices that specifically bind to the tag. Subsequently, proteins are trypsinized and analysed via mass spectrometry. (E) After trypsin digestion, the C‐terminal fragments of mammalian SUMO family members are too large to efficiently map the SUMO‐conjugated lysines in target proteins. To enable site‐specific purification, protease cleavage sites are introduced in the C‐termini of mammalian SUMO family members. SUMO target proteins modified with these mutant versions of SUMOs are fused to specific protein tags and purified as previously described for epitope or affinity tags. Subsequent digestion with either trypsin or the endoproteinase LysC, depending on the SUMO mutant employed, results in shorter SUMO peptides to facilitate identification of SUMO sites via mass spectrometry. (F) Alignment of the C‐termini of mature SMT3 from yeast and mature human ubiquitin, SUMO1, SUMO2 and SUMO3, demonstrating the various lengths of the tryptic remnants remaining after cleavage. Arginine and lysine residues are highlighted in red. The mutations used to facilitate identification of SUMO2 sites are indicated by arrows.
