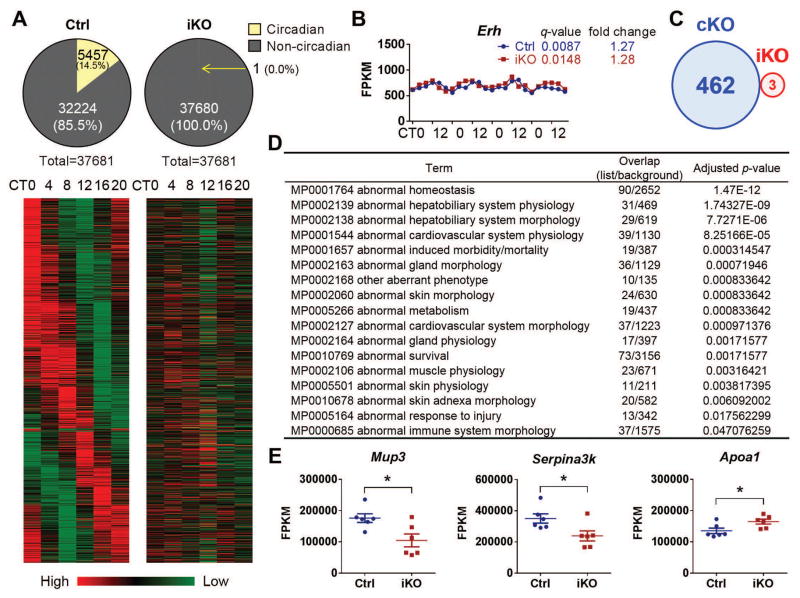
Fig. 6. Hepatic transcriptome.
RNA samples from iKOs and their littermates under DD condition were used for RNA-Seq. (A) Out of the 37,681 transcripts, 5,457 exhibited circadian variability in Ctrl mice. By contrast, only 1 gene in iKOs exhibited a circadian pattern. Heat map rendering of the temporal gene expression pattern of these 5,457 circadian genes is shown with the average gene expression level measured for each time point. (B) Erh is the only hepatic gene that shows a circadian expression pattern in iKOs, and the q-value (JTK_CYCLE analysis) and fold change (ratio of peak/trough) are shown [x-axis, circadian time; y-axis, fragments per kilobase of transcript per million mapped reads (FPKM)]. (C) A Venn diagram (sizes not to scale) depicting the number of differentially expressed genes in cKO and iKO strains. (D) MGI Mammalian Phenotype Level 3 enrichment analysis for cKO differentially expressed genes. The top 17 (adjusted P value<0.05) phenotypes related to these genes are given. Overlap, the number of appearing gene/the number of background genes. (E) Mup3, Serpina3k, and Apoa1 are the only differentially expressed genes in the iKO strain.