Abstract
Plant disease resistance occurs as a hypersensitive response (HR) at the site of attempted pathogen invasion. This specific event is initiated in response to recognition of pathogen-associated molecular pattern (PAMP) and subsequent PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI). Both PTI and ETI mechanisms are tightly connected with reactive oxygen species (ROS) production and disease resistance that involves distinct biphasic ROS production as one of its pivotal plant immune responses. This unique oxidative burst is strongly dependent on the resistant cultivars because a monophasic ROS burst is a hallmark of the susceptible cultivars. However, the cause of the differential ROS burst remains unknown. In the study here, we revealed the plausible underlying mechanism of the differential ROS burst through functional understanding of the Magnaporthe oryzae (M. oryzae) AVR effector, AVR-Pii. We performed yeast two-hybrid (Y2H) screening using AVR-Pii as bait and isolated rice NADP-malic enzyme2 (Os-NADP-ME2) as the rice target protein. To our surprise, deletion of the rice Os-NADP-ME2 gene in a resistant rice cultivar disrupted innate immunity against the rice blast fungus. Malic enzyme activity and inhibition studies demonstrated that AVR-Pii proteins specifically inhibit in vitro NADP-ME activity. Overall, we demonstrate that rice blast fungus, M. oryzae attenuates the host ROS burst via AVR-Pii-mediated inhibition of Os-NADP-ME2, which is indispensable in ROS metabolism for the innate immunity of rice. This characterization of the regulation of the host oxidative burst will help to elucidate how the products of AVR genes function associated with virulence of the pathogen.
Keywords: AVR-effectors, gene-for-gene interaction, NADP-Malic enzyme, reactive oxygen species, rice
INTRODUCTION
Many scientists have striven to find ways to control hemibiotrophic M. oryzae, a well-characterized fungal pathogen that causes rice blast disease. However, this devastating disease is occurring in other crops, thus attracting more attention from the scientific community (Maciel et al., 2014). This issue is associated with a global food shortage and security (Fisher et al., 2012), thereby leading many businesses and scientists to attempt to pinpoint the detail mechanism of the pathogenesis (Giraldo et al., 2013). Significant advances have been made as regards genetic evidence (Liu et al., 2010; Zhang et al., 2015) but the underlying issue remains to be solved to ensure crop protection. Here, we aimed to explore a novel strategy to control rice blast disease as a model system of major hemibiotrophic fungal pathogens through elucidation of a virulence mechanism of pathogen’s AVR effector.
Pathogen AVR effectors have been genetically proven to be essential components in plant immune responses (Flor, 1971). However, the mechanism by which AVR effectors and R proteins are associated with these responses remains unclear. Initially, the ligand-receptor model (Gabriel and Rolfe, 1990) was widely supported, but the lack of physical interactions between a number of R/AVR pairs has resulted in the generation of alternative guard and decoy hypothesis (McHale et al., 2006; van der Hoorn and Kamoun, 2008). One of the interesting results have been reported recently for AVR-Pii and OsExo70-F3 interaction in which OsExo70-F3 physically interacts with AVR-Pii and specifically involved in Pii-dependent resistance suggesting OsExo70-F3 as a helper in Pii/AVR-Pii interactions (Fujisaki et al., 2015). Physical interactions that underlie R/AVR function are starting to be elucidated but still largely ambiguous (Cesari et al., 2013; 2014a; 2014b; Maqbool et al., 2015; Williams et al., 2014). More recently, rice resistant protein pair RGA4/RGA5, was shown to be required for recognition of Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 revealing a mode of AVR protein recognition through direct binding to a novel, non-LRR interaction domain (Cesari et al., 2013; 2014b). In bacterial pathogens, with the identification of type III secretion systems, effector proteins were demonstrated to be crucial for virulence (Dean, 2011). In fungal pathogens, the biological activities of effectors have only recently been characterized as virulence factors, as well as signals to trigger host defense responses (Dangl et al., 2013; Giraldo and Valent, 2013).
Once pathogen effectors are secreted into the host cytoplasm, they induce a reprogramming of host metabolomes (Parker et al., 2009) and transcriptomes (Wei et al., 2013). However, many effectors only partially contribute to virulence except for a few critical effectors known as “core effectors” (Dangl et al., 2013). Some AVR effectors represent typical core effectors and are defined by their substantial contribution to suppression of the plant immune system (Fujisaki et al., 2015; Mackey et al., 2003; Park et al., 2012). In addition to the benefits of AVR effectors, they are also detrimental to the pathogen itself because the detection of AVR effectors by R proteins can halt pathogen proliferation by rapid HR cell death (Greenberg and Yao, 2004). Thus, it will be interesting to reveal the evolutionary and functional advantages of AVR genes regardless of their distinct disadvantages.
After a plant senses a pathogen signal, the response leads to activated PTI and subsequent ETI involving a rapid ROS burst (Stael et al., 2015). Scientists have revealed the pivotal role of ROS in the infection response (Levine et al., 1994; Pogány et al., 2009; Torres et al., 2005; 2006). As exemplified by the essential role of the Ustilago maydis effector Pep1, an inhibitor of peroxidases, for all host cell penetration events fast suppression of host ROS production is vital to establish compatibility (Doehlemann et al., 2009; Hemetsberger et al., 2012). Furthermore, the mechanism of distinct biphasic ROS accumulation has been demonstrated in a diverse array of avirulent plant-pathogen interactions involving bacteria, fungi, and viruses (Able, 2003; Doke, 1983; Grant and Loake, 2000; Piedras et al., 1998). However, these studies could not demonstrate how the biphasic ROS production mechanism is associated only with incompatible interactions. Therefore, we aimed to unveil the plausible cause of active suppression of ROS burst in compatible interactions with the help of M. oryzae AVR-Pii-Os-NADP-ME2-3 interactions.
The nicotinamide adenine dinucleotide phosphate (NADPH) oxidase D (NADPH-oxidase D) is a plasma membrane-bound enzyme that generates ROS (Kadota et al., 2015) using the cytosolic electron donor NADPH, supplied by NADP-ME2. NADP-ME2 is a rapidly and abundantly induced enzyme associated with ROS production following M. oryzae infection (Parker et al., 2009; Wei et al., 2013). Notably, NADP-ME2 activity increases rapidly only in resistant cultivars; however, it is significantly suppressed in susceptible cultivars (Parker et al., 2009). This virulent race-specific NADP-ME2 suppression is strongly suggestive of NADP-ME2 involvement in plant disease resistance.
Here, we provide functional evidence that M. oryzae AVR-Pii, which interacts with rice Os-NADP-ME2, attenuates the ROS burst and plays an essential role as one of the key pathogenicity determinants in disrupting host innate immunity.
MATERIALS AND METHODS
Plant materials and growth conditions
Rice cultivars Hwayeongbye (HY), Nipponbare (NB), and the ΔOs-nadp-me2-3 mutant were used in all experiments. ΔOs-nadp-me2-3 T-DNA insertion mutant seeds were provided by Rice Functional Genomic Express Database (RiceGE) managed by the Salk Institute. (http://signal.salk.edu./cgi-bin/RiceGE) (Jeon et al., 2000). Plants were placed in growth chambers under 60% humidity and 16 h light and 8 h dark conditions at 28°C. All WT rice seeds were obtained from the National Institute of Crop Science (http://www.nics.go.kr).
Fungal culture conditions and transformation
The fungal strain M. oryzae INA168 was obtained from the Center for Fungal Genetic Resources (Seoul National University, Seoul, Korea; http://genebank.snu.ac.kr). All fungal strains were stored at −20°C and normally grown on rice bran agar media (20 g rice bran, 20 g sugar, and 20 g agar in 1000 ml Milli-Q water). M. oryzae INA168 was grown under continuous light conditions for 10–12 days at 25°C for spore production. A hygromycin-resistant mCherry-tagged AVR-Pii transformant was produced by polyethylene glycol (PEG)-mediated protoplast transformation, as described previously with minor modifications (Jeon et al., 2008). M. oryzae INA168 mycelia grown in liquid culture for 3 days were incubated with 5 mg/ml lysing enzyme (Sigma-Aldrich, L1412) for 3–4 h using a shaker at 9–12 rpm. Ten micrograms of BAS4:EGFP and ProAVR-Pii:AVR-Pii: mCherry (Fig. 4A) and pSK1044 (Park et al., 2013) were transformed into freshly harvested protoplasts (1 × 107/ml). The positive transformants were detected on TB3 agar media using hygromycin selection (20 g sucrose, 0.3 g yeast extract, 0.3 g casamino acids, 1 g glucose, 0.8 g agar, and 0.2 mg/ml hygromycin in 100 ml) and confirmed by PCR using Os-NADP-ME2-3-specific and hygromycin-specific primers (Supplementary Table S4). The yeast strain MAV203 was used for Y2H screening. MAV203 was cultured normally in yeast extract peptone dextrose (YPD) solid agar media (10 g yeast extract, 20 g bacto peptone, 20 g agar powder, 40 ml 50% glucose in 1l volume) at 30°C for 3 days.
Fig. 4.
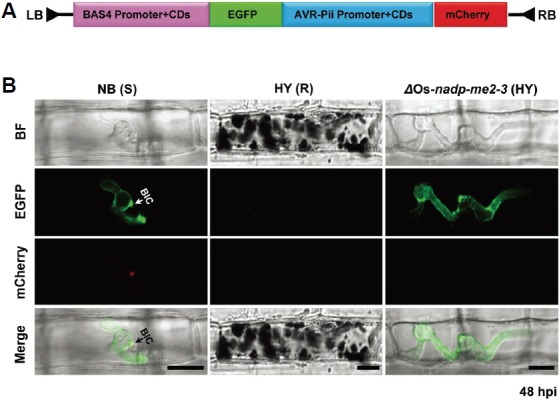
Differential regulation of the effectors was visualized using two BAS4 and AVR-Pii-effector expression cassettes. (A) Schematic representation of plasmid pCB1004 containing BAS4:EGFP and AVR-Pii:mCherry as described in the “Materials and Methods”. (B) Expression of BAS4 and AVR-Pii was observed in susceptible NB, where BAS4 appeared uniformly as a bright outline in the invasive hyphae, and AVR-Pii accumulated locally in the BIC (white arrow) at 24-48 hpi. In contrast, expression of BAS4 appeared in invasive hyphae, but AVR-Pii was not detected in the ΔOs-NADP-ME2-3 mutant. In addition, BAS4 and AVR-Pii were not observed in HY, indicating no viable hyphae. BIC, biotrophic interfacial complex; S, susceptible; R, resistant. Scale bars, 10 μm.
Quantitative Real-time PCR (qPCR)
To analyze the transcriptional level of Os-NADP-ME2-3 in ΔOs-nadp-me2-3 mutant lines, qPCR (Stratagene Mx3000P) was performed. Total RNA was isolated from rice tissues using TRIzol reagent and quantified by the spectrophotometric method. First-strand cDNA was synthesized in a 20 μl reaction mixture using a cDNA synthesis kit (Invitrogen) according to the protocol provided by the manufacturer. Furthermore, 1 μl prepared cDNA was used as a template for reverse transcription (RT)-PCR and qPCR. qPCR was performed using Brilliant III Ultra-Fast SYBR Green PCR Master Mix (Stratagene) according to the manufacturer’s protocol. Actin and ubiquitin were used as controls for this study. The list of primers used in this experiment is provided in Supplementary Table S4.
Rice blast inoculations
For the spray inoculation method, incompatible M. oryzae race, INA168 spores were harvested from 10–12-day culture plates using Milli-Q water with 0.025% of Tween 20 (Sigma-Aldrich, P2287). The spore concentration was adjusted to 4 × 105 spores/ml. The harvested spore suspension was sprayed over 2-week-old rice seedlings (HY, NB, and ΔOs-nadp-me2-3 mutant). The inoculated seedlings were incubated at 25–28°C for 24 h under dark conditions. After 24 h, the inoculated plants were moved to normal conditions (16 h/8 h dark/light), as described previously (Kim et al., 2009). Disease phenotype was observed at 4 dpi.
Cloning and plasmid construction
The coding regions of four alternative splice variants of Os-NADP-ME2-3 (LOC_Os01g52500) were amplified using the rice cDNA library and primers containing Gateway attB1 and attB2 sites (Supplementary Table S4). Primers were designed using the information provided by the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). Using the same approach, AVR-Pii cDNA was amplified from M. oryzae INA168 gDNA, using specific primers with and without signal peptides (Supplementary Table S4). The amplified coding regions of Os-NADP-ME2-3 and AVR-Pii were subcloned into the pDONRTM201 entry vector using BP clonase (Invitrogen) to create entry clones. The entry clones were recombined into Gateway destination vectors pDEST32 and pDEST22 using the Gateway system (AVR-Pii:pDEST32; Os-NADP-ME2-3:pDEST22) to generate bait and prey, respectively, for Y2H screening, C-terminal Venus (pDEST-VYCE®GW; bait) and N-terminal Venus (pDEST-VYNE®GW; prey) for BiFC (Singh et al., 2012; 2014a; 2014b). In addition, the AVR-Pii mutant (AVR-Pii-MT) was amplified from AVR-Pii-MT:pGEX4T-1 using AVR-Pii-specific primers containing the attB1 and attB2 sites. The final construct (AVR-Pii-MT : pDEST32) was generated as described above for the Y2H assay. For protein expression, the AVR-Pii open reading frame (ORF) was subcloned into pBlueScript II SK (pBSK) using BamHI and NotI adapter sites and transferred into N-terminal glutathione S-transferase (GST)-tagged pGEX4T-1 (protein expression vector). In the same manner, the ORFs of Os-NADP-ME2-3 and Os-NADP-ME2-2 were subcloned into pBSK with NdeI and XhoI adaptor sites and transferred to N-terminal IF2 and 6xHis-tagged pET28a (protein expression vector). The final clones of both genes were confirmed by nucleotide sequencing (Macrogene).
To construct the AVR-Pii:mCherry expression cassette, mCherry was cloned into pCB1004 (Yoshida et al., 2009) with XbaI and SacI adapter sites. Further, the AVR-Pii ORF, with a 543 bp promoter region, was amplified from M. oryzae INA168 gDNA using AVR-Pii promoter forward-1 and AVR-Pii w-sp reverse primers with KpnI and XbaI adapter sites. The PCR-amplified cDNA fragment was cloned into pBSK and inserted in front of mCherry in pCB1004, generating the 543 bp ProAVR-Pii: AVR-Pii:mCherry:pCB1004. The upstream 404 bp promoter was cloned into pBSK and transferred into SalI and EcoRV sites in front of a 543 bp promoter in ProAVR-Pii:AVR-Pii: mCherry:pCB1004. The effector BAS4:EGFP expression cassette was PCR-amplified from pBV324 (Mosquera et al., 2009) using BAS4-EGFP forward and reverse primers with the adapter enzyme site SalI, and subcloned into pBSK. The BAS4:EGFP cassette was cloned in front of the 947 bp ProAVR-Pii:AVR-Pii:mCherry cassette in pCB1004, generating the BAS4:EGFP and ProAVR-Pii:AVR-Pii:mCherry:pCB1004 vector (Fig. 4A). Lastly, pBV551 (Giraldo et al., 2013) containing ProPWL2:PWL2 and BAS4:EGFP cassette was kindly provided by the Center for Fungal Genetic Resources (Seoul National University, Seoul, Korea; http://genebank.snu.ac.kr). All primer information is provided in Supplementary Table S4.
Inverse PCR for native AVR-Pii promoter
Inverse PCR was performed using two different sets of AVR-Pii promoter upstream forward and reverse primers for each restriction enzyme. The newly identified AVR-Pii promoter sequences were reconfirmed by direct sequencing of the M. oryzae INA168 gDNA, using two primers (upstream-1346-forward and upstream-2-outer reverse). The entire 1.6 kb promoter was identified and used further as a template to amplify an additional 404 bp of the promoter using AVR-Pii promoter forward-2 and AVR-Pii promoter upstream-1 inner reverse primers (Supplementary Fig. S8). All primer information is provided in Supplementary Table S4.
Site-directed mutagenesis
The AVR-Pii mutant (AVR-Pii-MT) was obtained by mutation of five negatively charged amino acids (Supplementary Figs. S3A and S3B) using the AVR-Pii:pGEX4T-1 plasmid, as a template. Glutamate at position 23 and aspartate at positions 27, 41, 42 and 44 in AVR-Pii cDNA (without the signal peptide) were mutated to alanine. Site-directed mutagenesis of AVR-Pii was performed using the QuikChange® Site-Directed Mutagenesis Kit from Stratagene. AVR-Pii-MT1 forward and reverse primers with D41A, D42A and D44A were used for the first PCR. After PCR and PCR-purification, the methylated, non-mutated and parental DNAs were digested using DpnI, and the newly synthesized site-directed mutagenized plasmids were transformed into the E. coli strain TOP10. The required mutated fragment was obtained by overlap PCR using pGEX5 and pGEX3 primers. Finally, the fragment was ligated into pGEX4T-1 and named AVR-Pii-MT1:pGEX4T-1. The recovered mutated clone was confirmed by nucleotide sequencing (Cosmo-GenTech). AVR-Pii-MT1:pGEX4T-1 was used as a template to obtain additional E23A and D27A mutations using two new AVR-Pii-MT forward and reverse oligos. A procedure similar to the one described above was applied to obtain the final GST-tagged mutated AVR-Pii cDNA, which was named AVR-Pii-MT:pGEX4T-1. All primers are listed in Supplementary Table S4.
Yeast two-hybrid assay
The Y2H screening of M. oryzae AVR-Pii without signal peptides using the rice cDNA library was performed as described previously (Singh et al., 2012; 2014a). The Mav203 transformed bait (AVR-Pii) and prey (cDNA library) interaction was confirmed using X-gal assays, and the growth pattern in standard selection media lacking tryptophan, leucine, and histidine, supplemented with 55 mM 3-AT (SC-LTH; 6.7 g YNB with ammonium sulfate, 0.65 g CSM-LEU-TRP-HIS, 20 g agar, 40 ml 50% glucose and 55 mM of 3-AT/L), medium lacking uracil (SC-URA; 6.7 g YNB with ammonium sulfate, 0.64 g CSM-URA, 20 g agar, 40 ml 50% glucose/L), medium lacking leucine and tryptophan (SC-LT; 6.7 g YNB with ammonium sulfate, 0.64 g CSM-LEU-TRP, 20 g agar, 40 ml 50% glucose/L), and a lack of growth in medium containing 5-FOA (0.2%) (SC-LT+ 0.2% 5-FOA; 6.7 g YNB with ammonium sulfate, 0.64 g CSM-LEU-TRP, 20 g agar, 40 ml 50% glucose/L) as described previously (Singh et al., 2012). Strong interactor cells were observed as blue on YPD plates within 24 h after the X-gal assay and grew within 4 d on SC-LTH, SC-URA and SC-LT, but not on SC-LT with 0.2% 5-FOA (SC-FOA). Similarly, other AVR-effectors (AVR-Pia, AVR-Pizt and AVR-Pikm) were used as bait and Os-NADP-ME2-3 as the prey protein in the X-gal assay. Strong (pEXP32/Krev1+pEXP22/RalGDS-wt), weak (pEXP32/Krev1+pEXP22/RalGDS-m1), and absent (pEXP32/Krev1+ pEXP22/RalGDS-m2) were used as controls, as purchased from Invitrogen (ProQuest Two-Hybrid System). Krev1 is a member of the Ras family of GTP-binding proteins, and RalGDS is the Ral guanine nucleotide dissociator stimulator protein. RalGDS-m1 and RalGDS-m2 are mutants of RalGDS that affect its interaction with Krev1. In addition, to avoid false positives, autoactivation of bait/prey, bait/prey swapping and retransformed assays were done as described previously (Singh et al., 2012; 2014a).
Bimolecular fluorescence complementation (BiFC)
Rice leaf sheath preparation, onion tissue preparation, DNA preparation, co-transformation, and biolistic bombardment were performed as described previously (Singh et al., 2012). To detect interactions, 20 μg bait protein (AVR-Pii:pDEST-VYCE®GW) and prey protein (Os-NADP-ME2-3:pDEST-VYNE ®GW) were mixed and bombarded in onion and rice epidermal cells, using biolistic bombardment (Bio-Rad, Biolistic ®-PDS-1000/He Particle Delivery System), as described previously (Singh et al., 2012). They were incubated at 25°C for 12-24 h in the dark followed by cell imaging using a confocal laser microscope (Leica, TCS SP5) at 20× (onion cells) and 40× (rice cells) with a Venus filter (Ex/Em: 488 nm/505-550 nm wavelength). In addition, for the negative control, 20 μg of each AVR-Pii:pDEST-VYCE®GW and pDEST-VYNE®GW; Os-NADP-ME2-3:pDEST-VYNE®GW and pDEST-VYCE®GW; pDEST-VYCE®GW and pDEST-VYNE®GW were bombarded in rice and onion epidermal cells. Similarly, Arabidopsis pEXP-VYCE(R)-cnx7 and pEXP-VYNE(R)-cnx6 were used as a positive control. The image was obtained as explained by in vivo live-cell imaging.
Co-immunoprecipitation
The 35Sp:4xMyc:AVR-Pii (bait) and 35Sp:3xHA:Os-NADP-ME2-3 (prey) plasmids were generated using the 4xMyc-tagged Co-IP vector pGWB18 and 3xHA-tagged pGWB15, as described previously (Singh et al., 2012). The transient expression assay was performed by infiltrating tobacco leaves with Agrobacterium tumefaciens cells (strain C58C1 pCH32) with bait, prey and p19 as explained earlier (Singh et al., 2012). The samples were collected after 3 days of incubation, and fine powder was produced using liquid nitrogen. The protein was extracted from each sample using Co-IP buffer (10% Glycerol, 25 mM Tris-CL(pH 7.5), 1 mM EDTA, 150 mM NaCl, 1% Triton X-100 and 1X protease inhibitor cocktail). For Co-IP, 500 μg each protein was incubated with 20 μl proteinase A sepharose beads (Amersham Biosciences) at 4°C for 2 h. Immunoprecipitation was performed using the monoclonal antibody Myc, followed by pre-coupling with proteinase A sepharose beads. The protein complex was further washed three to four times with 1X PBS (phosphate-saline buffer), and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE 8%) was performed. The proteins were then blotted with the monoclonal antibody HA (1:1000; ABM Inc., GO36) and Myc (1:1000; ABM Inc., GO19).
In vitro pull-down assay
The N-terminal glutathione S-transferase (GST)-tagged AVR-Pii and AVR-Pii-MT, N-terminal IF2, and 6xHis-tagged Os-NADP-ME2-3 and Os-NADP-ME2-2 were expressed in E. coli strain Rosetta followed by protein expression at 18°C for 16 h, 37°C for 4 h, 15°C for 5 h and 26°C for 5 h with 0.2 mM IPTG, respectively. GST alone, GST:AVR-Pii and GST:AVR-Pii-MT were purified using Glutathione-Sepharose 4B (Amersham Biosciences), and 6xHis:Os-NADP-ME2-3 and 6xHis:Os-NADP-ME2-2 were purified by Nickel affinity chromatography (Supplementary Figs. S9 and S10). The GST pull-down assay was performed as described previously (Singh et al., 2012). In brief, 5 μg purified GST:AVR-Pii was adsorbed onto 40 μl Glutathione-Sepharose 4B (GST beads), followed by the addition of 5 μg 6xHis:Os-NADP-ME2-3 and 6xHis:Os-NADP-ME2-2. A similar procedure was applied using 5 μg GST:AVR-Pii-MT and GST alone with 6xHis:Os-NADP-ME2-3. After incubation at 4°C for 4–12 h, unbound proteins were washed away using 1X PBS. The bound proteins were separated by 8% SDS-PAGE and visualized by coomassie brilliant blue (CBB) staining. The bound Os-NADP-ME2-3 and Os-NADP-ME2-2 were detected by immunoblotting using an anti-His tag antibody (1:1000; ABM Inc., GO20).
In vivo live-cell imaging and quantification of infected cells
Rice varieties HY, NB, and ΔOs-nadp-me2-3 were grown in a rice growth chamber for 4–5 weeks. The sheath inoculation method was performed as described previously, with minor modifications (Kankanala et al., 2007). Middle-aged leaf sheaths of each rice cultivar were cut into strips ∼7 cm long, which were inoculated by freshly harvested spores of M. oryzae INA168 at 4 × 105/ml concentration. The spore inoculant was introduced into the hollow space (above the mid vein) of rice sheaths using a pipette. The inoculated sheaths were incubated in a moistened box at 25°C under dark conditions for 48 and 72 h. After incubation, the sheaths were trimmed to remove all green parts. The remaining middle thin epidermal layer was fixed on the slide and used for microscopic analysis. For in vivo imaging of the hyphal colonization, the M. oryzae INA168 transformant with pSK1044 (Park et al., 2013), as well as BAS4: EGFP and ProAVR-Pii:AVR-Pii:mCherry (Fig. 4A), were cultured for 10–12 days under continuous light. The spores were adjusted to a 4 × 105/ml concentration and inoculated into HY, NB, and ΔOs-nadp-me2-3 mutants as described above. All microscopic images were obtained using a 40× oil lens under a bright field, EGFP (Ex/Em 488/507 nm) and mCherry (Ex/Em 587/610 nm) filters in a confocal microscope (Leica, TCS SP5). At the same time, all infected cells were counted in each rice cultivar inoculated with GFP-tagged INA168. The infected cells were divided into two phenotypes: cells with viable IHs and cells with dead hyphae/hypersensitive response (HR). The counting method was repeated five times.
NADP-ME activity measurements and inhibition assays
NADP-ME activity was assayed spectrophotometrically by monitoring NADPH production at 340 nm, using a standard reaction mixture (Detarsio et al., 2004) containing 67 mM Tris-Cl (pH 7.4), 3.5 mM L-malic acid, 0.3 mM NADP+, 5 mM MnCl2, 20 nM Os-NADP-ME2-3 and Os-NADP-ME2-2 in a final volume of 0.7 ml. The reaction was initiated by adding thrombin-treated Os-NADP-ME2-3 and Os-NADP-ME2-2. Initial velocity studies were performed by varying the concentration of one substrate around its Km value, while keeping the other substrate concentration at its saturation level. Kinetic parameters were determined by non-linear regression analysis, and data were fitted to the Michaelis-Menten equation using the curvefitter program of SigmaPlot 10.0 (Erkraft, Germany). Inhibition studies were performed similarly, using 10 nM AVR-Pii and AVR-Pii-MT as inhibitors. The initial velocity studies were performed by varying the concentration of L-malate (0.5-4 mM), while keeping the concentration of NADP+ at its saturation level (0.3 mM) and vice versa. The reaction was started by addition of 20 nM of OsNADP-ME2-3 and 10 nM of AVR-Pii. The competitive inhibition was determined by a double reciprocal plot between 1/Vo and 1/[S]. The dissociation constant of the inhibitors (Ki) was a calculated gradient or intercept term, obtained from the double reciprocal plot. The equation for competitive inhibition is given by
- Km
Michaelis constant in the absence of an inhibitor
- Vmax
maximum rate in the absence of an inhibitor
- [I]
Concentration of an inhibitor
- Ki
Dissociation constant of the inhibitors
Similarly, the inhibition assays of Os-NADP-ME2-3 by AVR-Pii-MT and Os-NADP-ME2-2 by AVR-Pii were determined by the same protocol. All activity assays were carried out at 25°C using a SP2000UV spectrophotometer (SmartPlus). The protein concentration was determined by the Bio-Rad Protein Assay using BSA as a standard.
Measurement of PAMP-triggered ROS production
ROS production was measured as described previously (Park et al., 2012) with minor modifications. ROS were elicited using chitin and sterile water as a negative control. Briefly, 16 leaf discs from 4 to 5-week-old rice cultivar HY and ΔOs-nadp-me2-3 were soaked in sterile distilled water overnight in 96-well plates. The next day, the water was replaced with 100 μl luminol (Bio-Rad Immun-star Horseraddish peroxidase substrate 170-5041), 1 μl horseradish peroxidase (Jackson Immunoresearch, 061-030-084), and 8 nM hexa-N-acetylchitohexaose (Santa cruz Biotechnology, Inc. sc-222018) and sterile water (as a control). Luminescence was measured after 10 min using a luminometer (SpectraMax L Microplate Reader). The standard deviation was calculated for each treatment.
Accession numbers
Sequence data from this article could be found on the Rice Genome Project website (http://rice.plantbiology.msu.edu/) or the EMBL/GenBank data libraries under accession numbers: Os-NADP-ME2-1, LOC_Os01g52500.1; Os-NADP-ME2-2, LOC_Os01g52500.2; Os-NADP-ME2-3, LOC_Os01g52500.3; Os-NADP-ME2-4, LOC_Os01g52500.4; Os-Exo70F2, LOC_Os02g30230; Os-Exo70F3, LOC_Os04g31330; Os-Exo70F4, LOC_Os08g41820; AVR-Pii, AB498874; BAS4: FJ807767; Hygromycin, AEJ60084.1; Actin, LOC_Os03g50885; Ubiquitin, LOC_Os06g46770.
RESULTS
Magnaporthe oryzae AVR-Pii specifically interacts with rice Os-NADP-ME2 in Yeast-two hybrid (Y2H) screening
We performed Y2H screen using M. oryzae AVR-effectors to identify rice host target components that undergo direct or indirect interactions to satisfy the current gene-for-gene interaction models (Gabriel and Rolfe, 1990; McHale et al., 2006; van der Hoorn and Kamoun, 2008). We identified several plant protein candidates for the AVR-Pii effector; one of them is Os-NADP-ME2 (Fig. 1A; Supplementary Table S1). Additional interactors for AVR-Pii effector were Os-Exo70F3, Os-Exo70F2 and Os-Exo70F4. AVR-Pii interaction with Os-Exo70F2 and Os-Exo70F3 was also reported by recent study in AVR-Pii-triggered immunity (Fujisaki et al., 2015). Detailed information of AVR-Pii effctor interactors were given in Supplementary Table S1. Because NADP-ME was previously shown to be important for immunity (Voll et al., 2012) and its suppression was observed only in compatible interactions (Parker et al., 2009), we focused on the interaction between AVR-Pii and NADP-ME. Thirteen different sizes of Os-NADP-ME2 cDNAs were screened using AVR-Pii bait based on Y2H screening (Fig. 1B). NAPD-ME2 produces NADPH during oxidative decarboxy-lation of malate into pyruvate in the presence of bivalent metal ions (Edwards and Andreo, 1992). In rice, four genes encoding NADP-MEs have been identified (Chi et al., 2004). Among these four genes, three are cytosolic isoforms (OsCytME1, OsCytME2, and OsCytME3) and one is a plastidic isoform (OsChlME) (Chi et al., 2004). The cytosolic isoform, OsCytME2, is referred to in our study as Os-NADP-ME2. We also found that the Os-NADP-ME2 gene produces five alternatively spliced mRNAs (Supplementary Fig. S1). Os-NADP-ME2-1 and Os-NADP-ME2-3 differ only in their C-terminus. Note that AVR-Pii only interacts with Os-NADP-ME2-1 and Os-NADP-ME2-3 out of five alternative isozymes (Fig. 1C). However, all recovered interacting clones belong to Os-NADP-ME2-3, which strongly suggests that only Os-NADP-ME2-3 is a unique target of AVR-Pii. The interaction-specificity between Os-NADP-ME2-3 and AVR-Pii was reconfirmed by screening each individual interaction against the other three AVR-effectors (AVR-Pia, AVR-Pizt, AVR-Pikm) (Fig. 1D), suggesting that each AVR-effector has a distinct interaction specificity with its own target proteins.
Fig. 1.
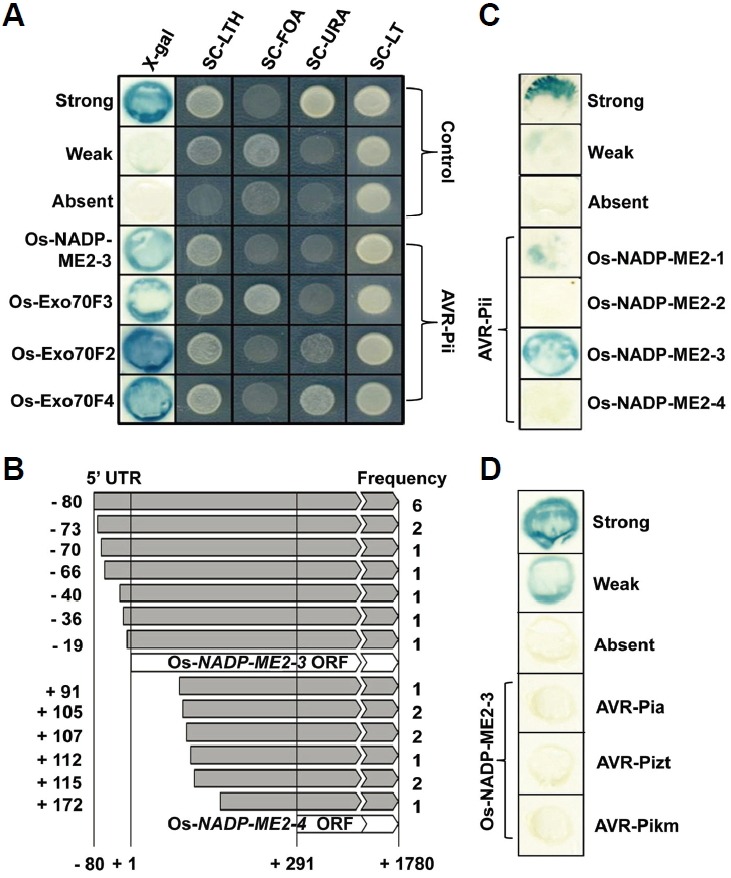
AVR-Pii preferentially interacts with Os-NADP-ME2-3. A-D, Interaction of AVR-Pii with Os-NADP-ME2-3 in Yeast-two hybrid (Y2H) assays. (A) Positive interactions were detected by five different tests, followed by the appearance of blue color in the X-gal assay and cell growth on synthetic complete-leucine-tryptophan-histidine (SC-LTH) including 55 mM 3-amino-1,2,4-triazole (3-AT), SC-leucine-tryptophan (SC-LT) containing 0.2% [w/v] 5-fluoroorotic acid (SC-FOA), SC-uracil (SC-URA) and SC-LT medias. Controls used were as follows: strong (pEXP32/Krev1 + pEXP22/RalGDS-wt); weak (pEXP32/Krev1 + pEXP22/RalGDS-m1); and absent (pEXP32/Krev1 + pEXP22/RalGDS-m2). (B) Thirteen different sizes of interacting Os-NADP-ME2-3 cDNAs with AVR-Pii were identified, and their detection frequencies are indicated. (C) AVR-Pii showed interaction specificity with the Os-NADP-ME2-1 and Os-NADP-ME2-3 alternative splice forms. (D) No interaction was detected between Os-NADP-ME2-3 and AVR-Pia, AVR-Pizt and AVR-Pikm.
Validation of interaction between AVR-Pii and Os-NADP-ME2-3
The interaction between Os-NADP-ME2-3 and AVR-Pii was further validated by bimolecular fluorescence complementation (BiFC) assay, Co-immunoprecipitation (Co-IP) and the glutathione S-transferase (GST) pull-down assay. The BiFC system is based on the formation of a fluorescent complex from the fusion of putative interacting proteins (Gehl et al., 2009). AVR-Pii and Os-NADP-ME2-3 were fused with the C-and N-terminal halves of Venus, then, subjected to the BiFC assay, along with Arabidopsis Cnx6 and Cnx7 as positive controls and empty vectors as negative controls. Fluorescence was observed in both interacting pairs and positive controls, whereas fluorescence was not found in the negative controls of rice (Oryza sativa) or onion (Allium cepa L.) cells (Fig. 2A; Supplementary Fig. S2). Co-IP was performed using 4xMyc-tagged AVR-Pii and 3xHA-tagged Os-NADP-ME2-3. Initially, we checked the transient expression of proteins by infiltrating it in tobacco (Nicotiana benthamiana) leaves as mentioned in Materials and Methods. Before performing Co-IP, the protein expression of 4xMyc-tagged AVR-Pii and 3xHA-tagged Os-NADP-ME2-3 were analyzed by immunoblotting with anti-Myc and anti-HA antibody respectively (Fig. 2B). After confirming the expression of AVR-Pii and Os-NADP-ME2-3, cell lysates were subjected to Co-IP using the anti-Myc antibody and immunoblotted using the anti-HA antibody (Fig. 2B). Next, AVR-Pii and Os-NADP-ME2-3 were expressed in Rosetta as GST and 6xHis-tagged recombinant proteins, followed by the in vitro pull-down assay (Fig. 2C). In addition, we generated mutated AVR-Pii (AVR-Pii-MT) by site-directed mutagenesis at five different sites (Supplementary Figs. S3A and S3B). Structural analysis of NADP-ME in maize (Zea mays L.) revealed that basic amino acids play key roles in catalysis (Detarsio et al., 2004); therefore, we randomly chose acidic residues in AVR-Pii and mutated them accordingly. The AVR-Pii-MT was tagged with a GST, followed by the pull-down assay using Os-NADP-ME2-3 (Fig. 2C). Similarly, the isoform Os-NADP-ME2-2 was tagged with 6xHis, and the pull-down assay was performed using AVR-Pii (Supplementary Fig. S4). The data suggested that AVR-Pii and Os-NADP-ME2-3 genuinely interact, while the change in structure of AVR-Pii disrupts this interaction, which was also validated by the Y2H system (Supplementary Fig. S5).
Fig. 2.
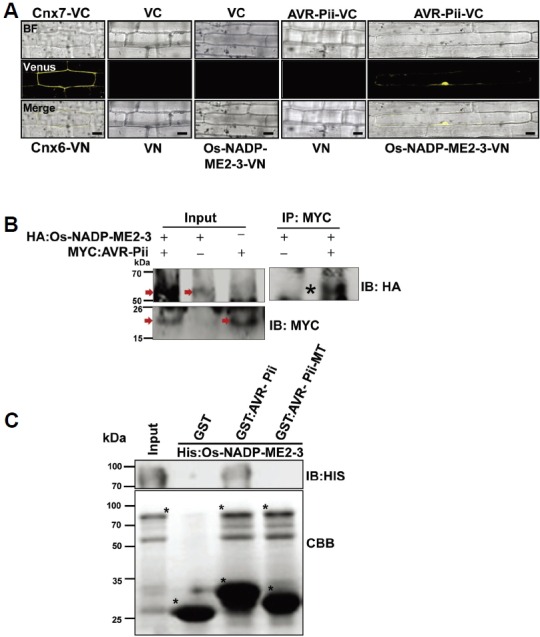
Validation of AVR-Pii and Os-NADP-ME2-3 interactions. (A) BiFC assay to validate the interaction of AVR-Pii (bait) and Os-NADP-ME2-3 (prey). Both bait and prey were fused to the C-and N-terminal halves of Venus and transformed to rice leaf sheaths using biolistic bombardment as described in the Materials and Methods section. The signal was detected using a Venus filter (Ex/Em: 488nm/505-550 nm wavelength) in a confocal microscope (Leica, TCS SP5). (B) AVR-Pii (without signal peptide) and Os-NADP-ME2-3 interaction was determined in planta by Co-IP. The protein extract was isolated from Nicotiana benthamiana leaves expressing the HA:Os-NADP-ME2-3 and Myc:AVR-Pii transgenes and protein expression was checked by anti-HA and anti-myc antibody respectively as shown by arrow in the Figure. The extract was immuno-precipitated by anti-Myc antibody and subjected to immunoblotting by anti-HA antibody. Asterisk indicates co-immuniprecipitated band of HA:Os-NADP-ME2-3 detected by anti-HA antibody. (C) In vitro GST pull-down assay of recombinant GST:AVR-Pii bound to rice 6xHis: Os-NADP-ME2-3. The pull-down and input samples were subjected to CBB staining as well as immunoblotting by the anti-His antibody (1:1000; ABM Inc., GO20). The result demonstrated that AVR-Pii genuinely interacts with Os-NADP-ME2-3, and mutated AVR-Pii (AVR-Pii-MT) fails to interact. The construction of AVR-Pii-MT is described in the Materials and Methods section. The asterisk (*) represents Histagged Os-NADP-ME2-3 (upper), GST alone, GST-tagged AVR-Pii and AVR-Pii-MT (lower). IP, Immunoprecipitation; IB, immunoblotting; CBB, Coomassie Brilliant Blue; VC, pDEST-VYCE®GW; VN, pDEST-VYNE®GW. Scale bars, 10 μm.
Deletion of the rice Os-NADP-ME2 gene disrupts innate immunity against the rice blast fungus
We found T-DNA insertion mutant rice from Rice Functional Genomic Express Database (RiceGE) developed by the Salk Institute. (http://signal.salk.edu./cgi-bin/RiceGE) (Jeon et al., 2000). The rice Os-NADP-ME2 gene was deleted (ΔOs-NADP-ME2-3) in the resistant rice cultivar Hwayeongbyeo (HY) by T-DNA insertional mutagenesis, but this did not affect any phenotype in terms of growth and yield under field conditions compared to the wild type (WT).This was similar to the phenotypic traits reported in Arabidopsis NADP-ME deletion mutants (Wheeler et al., 2005). T-DNA insertion was reconfirmed at the 5’-untranslated region (UTR) of Os-NADP-ME2 genomic DNA (gDNA) as described in the database (Fig. 3A). The genotype of the T3 progeny was analyzed using primers (RB+RP) and (S4+RP) to confirm transgene and homo/hetero selection, respectively (Fig. 3A; Supplementary Table S4). Inhibition of Os-NADP-ME2-3 transcription in ΔOs-nadp-me2-3 mutant lines was also confirmed using RT-PCR and quantitative real-time polymerase chain reaction (qPCR; Supplementary Figs. S6A and S6B). According to Yoshida et al. (Yoshida et al., 2009), resistant cultivars (R) show no or very small reddish hypersensitive responses (HR), while susceptible cultivars (S) show brown spindle-shaped necrotic lesions. In response to spray inoculation, the ΔOs-nadp-me2-3 mutant showed susceptible phenotypes to M. oryzae race INA168, whereas HY showed a resistant phenotype (Fig. 3B), demonstrating that deletion of OsNADDP-ME2-3 gene maintains virulence of INA168 towards ΔOs-nadp-me2-3 mutant.
Fig. 3.
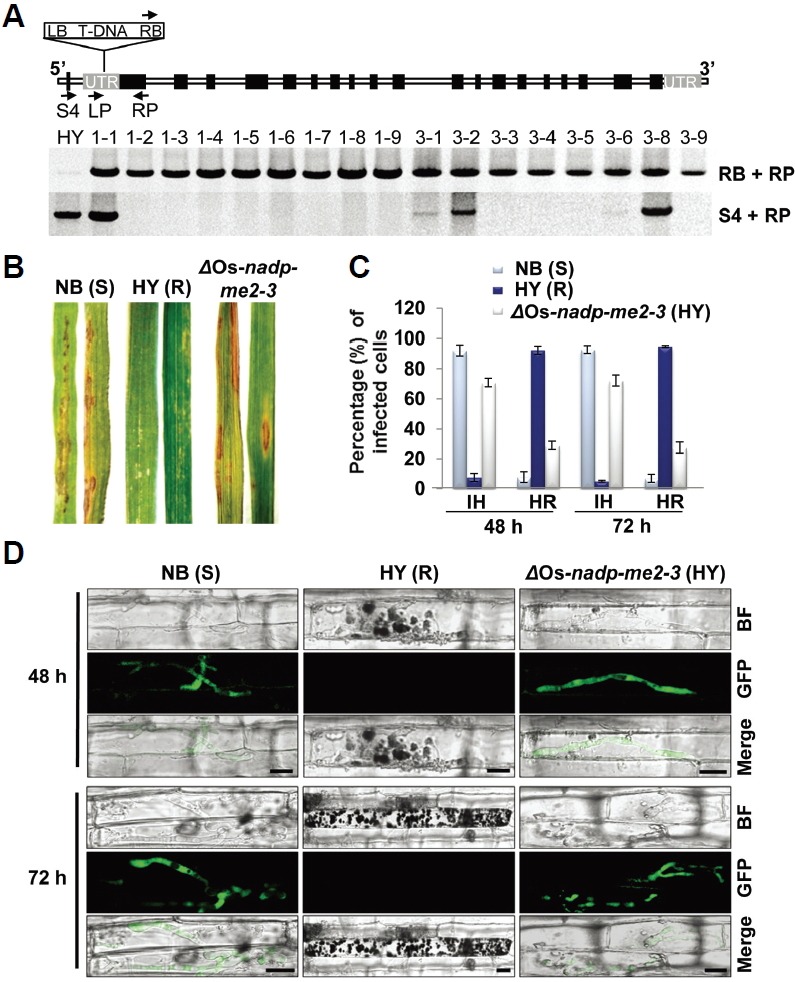
Deletion of Os-NADP-ME2-3 increases susceptible phenotype. (A) Genotyping of ΔOs-nadp-me2-3 mutant. The black arrows indicate the primer sets used for mutant confirmation and homo/hetero selection. RB, right border; LB, Left border. Primer pairs (RB + RP) and (S4 + RP) used for transgene analysis and homo/hetero selection respectively. (B) Rice seedlings of 2-week-old plants (HY, NB, and ΔOs-nadp-me2-3 mutant) were spray inoculated with the incompatible race INA168. The spore concentration was adjusted to 4 × 105 spores/ml. Leaves were photographed at 4 dpi. ΔOs-NADP-ME2-3 mutants showed susceptible phenotypes against M. oryzae INA168. NB, Nipponbare; HY, Hwayeongbyeo; S, susceptible; R, resistant. (C) Quantification of infected cells in HY, NB, and ΔOs-nadp-me2-3 mutants after inoculation with M. oryzae INA168 at 48 and 72 hpi. The error bars represent the standard deviation of five independent replicates. IH, Invasive hyphae; HR, Hyper sensitive response. (D) Live-cell imaging of M. oryzae infection of rice sheath epidermal cells. Successful colonization of GFP-tagged M. oryzae INA168 was observed in NB and ΔOs-nadp-me2-3 mutants, whereas all of the invasive hyphae lost their viability in HY. Scale bars, 10 μm.
Magnaporthe oryzae INA168 successfully colonizes in the susceptible NB cultivar and the ΔOs-nadp-me2-3 mutant but not in the resistant HY cultivar
To detect viable invasive hyphae during infection, we produced M. oryzae INA168 transformants that expressed the green fluorescent protein (GFP) by introducing the GFP tagged vector (Park et al., 2013). GFP-tagged transformant M. oryzae INA168 spore suspension was inoculated in rice epidermal cells of the susceptible cultivar NB, resistant cultivar HY and the ΔOs-nadp-me2-3 mutant. The viable colonizing hyphae of M. oryzae INA168 were observed in infected cells of susceptible NB and the ΔOs-nadp-me2-3 mutant, including neighboring cells, at 48 and 72 hour post inoculation (hpi) (Fig. 3D). In contrast to susceptible NB and the ΔOs-nadp-me2-3 mutant, all of the fungal hyphae detected within the infected cells of resistant HY lost viability with increasing infection time (Fig. 3D). Upon quantifying the infected cells between HY and ΔOs-nadp-me2-3 mutants, around 70% of the infected cells showed resistancebreakdown phenotype in ΔOs-nadp-me2-3 mutant compared to wild-type HY (Fig. 3C). These data are similar to recently reported data, which showed that the loss of cytosolic NADP-ME2 in Arabidopsis is associated with enhanced susceptibility to the hemibiotrophic fungal pathogen C. higginsianum (Voll et al., 2012). Hence, our data provide convincing evidence that M. oryzae INA168 recovered pathogenicity in the resistant cultivar HY after deletion of the Os-NADP-ME2 gene.
We also observed that the entire invaded HY sheath did not experience cell death after infection with M. oryzae race INA168. Many infected cells show viable phenotypes, likely due to a rapid host counterattack against fungal infection using apoplastic ROS bursts. Many attempted penetrations by M. oryzae leave remains of hyphal and appressorial debris on cell surface instead of causing host HR-mediated cell death (Supplementary Fig. S7). Similar NADPH oxidasederived suppression of cell death was observed in Arabidopsis (Torres et al., 2005). Cell death phenotypes were observed at 24–48 hpi in HY after M. oryzae race INA168 inoculation. Only heavily damaged cells or cells that require additional measures to stop further fungal IH might have adopted a second round of whole-cell death to minimize the damage within a single cell (Wang et al., 2009). This uncoupling ROS-derived resistance using host cell death/HR has been reported (Marino et al., 2012).
The AVR-Pii effector accumulates in the biotrophic interfacial complex (BIC)
AVR-effectors with native promoters preferentially accumulate in the BIC and translocate into the invaded cytoplasm, then move into neighboring cells, resulting in successful propagation of colonizing hyphae (Khang et al., 2010; Maqbool et al., 2015; Sharma et al., 2013). To analyze the expression pattern of the AVR-Pii effector, we constructed the mCherry-tagged AVR-Pii expression cassette under the control of a native promoter that was recovered by inverse PCR (Supplementary Fig. S8). We inserted the EGFP-tagged putative EHIM matrix protein biotrophy-associated secreted protein (BAS4:EGFP) in front of the AVR-Pii:mCherry cassette (Fig. 4A). BAS4 expression appears as a bright outline in the primary hyphae and inner layer of the BIC (Mosquera et al., 2009). The entire construction was then transformed into M. oryzae INA168 via PEG-mediated protoplast transformation. BAS4 expression appeared as uniform EGFP fluorescence in all invasive hyphae, and AVR-Pii: mCherry accumulated in the BIC in susceptible NB cultivar (Fig. 4B). These data suggested that AVR-Pii also accumulated in the BIC similarly to other M. oryzae AVR-effectors (Khang et al., 2010; Maqbool et al., 2015; Park et al., 2012; Sharma et al., 2013; Valent and Khang, 2010). In contrast to the response to susceptible NB cultivar, both fluorescences (BAS4:EGFP and AVR-Pii:mCherry) from transformant M. oryzae INA168 were lost in resistant HY cultivar, probably due to HR-induced cell death (Fig. 4B). However, normal BAS4:EGFP was observed, but accumulation of AVR-Pii was not detected in the ΔOs-NADP-ME2-3 mutant (Fig. 4). These data imply that AVR-Pii is regulated differentially from BAS4 during the biotrophic phase. In addition, PWL2, a BIC-translocated effector (Khang et al., 2010; Sharma et al., 2013), showed a similar expression pattern as AVR-Pii (Supplementary Fig. S9). The absence of PWL2 accumulation in ΔOs-nadp-me2-3 mutant suggested that the expression pattern of AVR-Pii in the ΔOs-nadp-me2-3 mutant is non-specific.
Purified AVR-Pii proteins inhibit in vitro NADP-ME activity
Malic enzyme activity and inhibition studies were performed using purified Os-NADP-ME2-3, Os-NADP-ME2-2, AVR-Pii and AVR-Pii-MT proteins (Supplementary Figs. S10 and S11). From an activity assay, the kinetic properties of both recombinant Os-NADP-ME2-3 and Os-NADP-ME2-2 were calculated. The maximum velocity (Vmax), turnover rate (Kcat) and catalytic efficiency (Kcat/Km) were higher for NADP+ than L-Malate in both cases, whereas these parameters were approximately two-fold higher for the reaction catalyzed by Os-NADP-ME2-3 than by Os-NADP-ME2-2 (Supplementary Table S2). The hyperbolic curve of Os-NADP-ME2-3 activity and its inhibition by AVR-Pii, as well as AVR-Pii-MT, are shown in Figs. 5A and 5B. There was a distinct difference in the activity of Os-NADP-ME2-3 in the absence and presence of the inhibitors (AVR-Pii-MT and AVR-Pii). Shortly after addition of AVR-Pii, the enzyme activity of Os-NADP-ME2-3 was decreased dramatically, while AVR-Pii-MT had no effect (Figs. 5A and 5B). This suggested that AVR-Pii inhibited the activity of Os-NADP-ME2-3, while mutation of AVR-Pii prevented this inhibition. The activity and inhibition data were applied to the Lineweaver-Burk equation (1/activity against 1/[substrate]). This data representation suggested that AVR-Pii competitively inhibited Os-NADP-ME2-3 activity (Fig. 6). Based on the double reciprocal plot, the dissociation constant (Ki) was calculated for both substrates. Ki for NADP+ was higher than that for L-Malate (Supplementary Table S3). Next, Os-NADP-ME2-2 showed extremely lower activity than that of Os-NADP-ME2-3. In addition, there was no inhibition of Os-NADP-ME2-2 by AVR-Pii (Supplementary Figs. S12 and S13). These results demonstrate that AVR-Pii but not AVR-Pii-MT specifically inhibits in vitro NADP-ME activity of OsNADP-ME2-3.
Fig. 5.
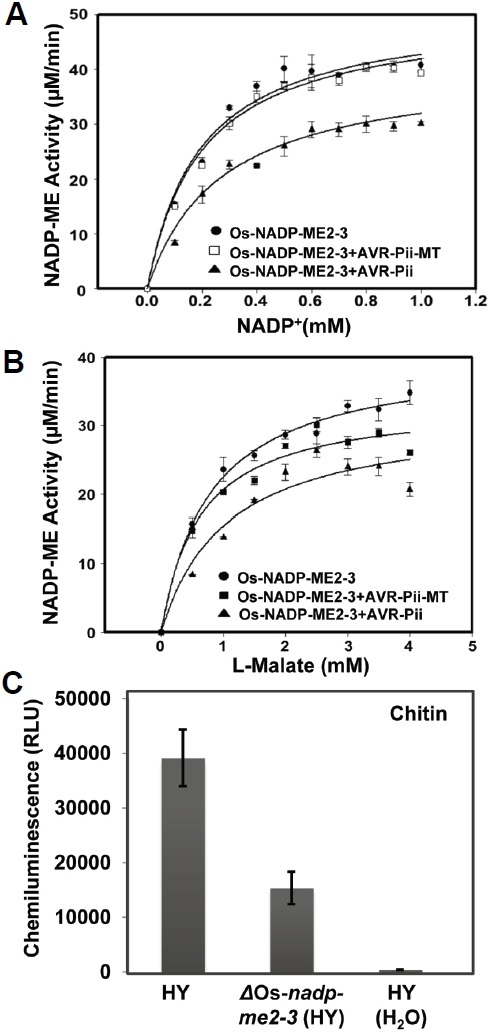
Inhibition of Os-NADP-ME2-3 attenuates PAMP-triggered ROS burst. (A) and (B), AVR-Pii specifically inhibits Os-NADP-ME2-3 activity. The activity of Os-NADP-ME2-3 alone or in the presence of AVR-Pii or AVR-Pii-MT, was determined at varying concentrations of NADP+ at a saturating concentration of L-Malate (3.5 mM) (A) and at varying concentrations of L-Malate at a saturating concentration of NADP+ (0.3 mM) (B) with fixed levels of other the substrates. Results are presented as mean ± SD of triplicate determinants. (C) Oxidative burst elicited by Chitin (8 nM) in HY and ΔOs-nadp-me2-3 leaf disc. Deletion of Os-NADP-ME2-3 inhibited PAMP-triggered ROS bursts. ROS generation was measured using luminol with a luminometer (SpectraMax L Microplate Reader). A total of 100 μl luminol, 1 μl horseradish peroxidase, and 8 nM chitin, and sterile water (as a control) were added to HY and ΔOs-nadp-me2-3 mutants. Values represent the mean ± SD (n = 16). The experiment was repeated three times with similar results.
Fig. 6.
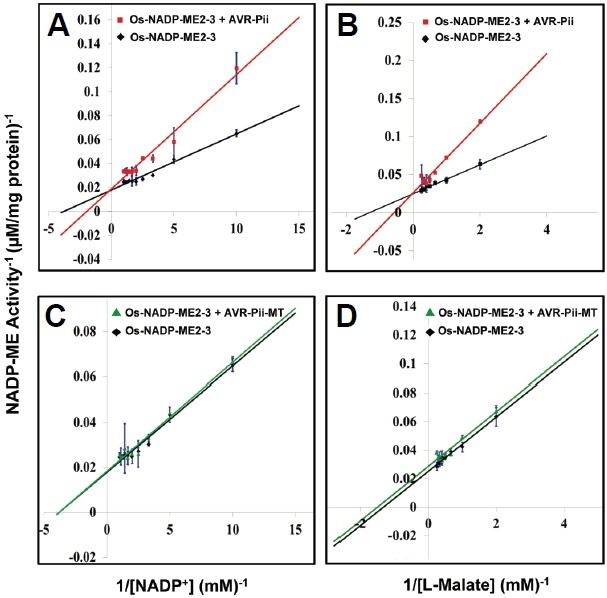
Double reciprocal plot for the inhibition of Os-NADP-ME2-3 by AVR-Pii and AVR-Pii-MT. The activity of Os-NADP-ME2-3 alone and in the presence of AVR-Pii or AVR-Pii-MT was determined using NADP+ as the variable substrate at 3.5 mM L-Malate (A) and (C) and using L-Malate as the variable substrate at 0.3 mM NADP+ (B) and (D). The data indicates AVR-Pii competitively inhibits Os-NADP-ME2-3, whereas a lack of inhibition was found with AVR-Pii-MT. The averages are from three independent repeats.
Os-NADP-ME2-3 is involved in PAMP-triggered ROS burst
Chitin is known fungal PAMP elicitor that generates ROS after its perception by plant cells. Rice cells can perceive chitin through pattern recognition receptor CEBiP/OsCERK1 (Shimizu et al., 2010). To determine whether ROS generation is affected in ΔOs-nadp-me2-3 mutants after PAMP elicitor treatment, we used leaves from wild-type HY and ΔOs-nadp-me2-3 mutant of HY background. ROS generation was measured using a luminol-based assay. The PAMP-triggered ROS burst was clearly observed in HY leaf discs, whereas ROS burst decreased by more than 50% in ΔOs-nadp-me2-3 mutant (Fig. 5C). This result suggests that Os-NADP-ME2-3 is directly involved in PAMP-triggered ROS bursts during the early infection process.
Our data provide evidence that AVR-Pii-triggered ROS inhibition (ATRI) helps M. oryzae INA168 to disrupt rice innate immunity and allows a successful biotrophic phase in rice cells after penetration (Fig. 7). This novel ATRI mechanism suggests that during infection the rice blast fungus M. oryzae attacks specific targets among countless host proteins. Mackey et al. (Mackey et al., 2003) also hypothesized that most pathogen virulence factors target essential machines of the host organism. Although rice ROS burst-assisted protection could not be applied to all plant pathogens, studies suggest that ROS production plays a fundamental role in HR-mediated resistance in plant-microbe interactions (Marino et al., 2012).
Fig. 7.
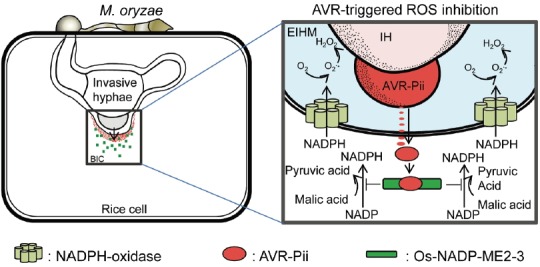
A model of AVR-Pii and Os-NADP-ME2-3 in M. oryzae–rice interactions. Specific binding affinity of AVR-Pii with Os-NADP-ME2-3 suppresses Os-NADP-ME2-3 and inhibits NADPH production activity near the BIC. In the absence of AVR-Pii, active Os-NADP-ME2-3 catalyzes malic acid into pyruvic acid, generating NADPH, and subsequent production of apoplastic ROS with increased resistance to M. oryzae. In AVR-triggered ROS inhibition, secretion of AVR-Pii helps to establish compatibility in susceptible rice cultivar. EIHM, extra-invasive hyphal membrane; IH, invasive hyphae.
DISCUSSION
Insights into the function of secreted fungal effectors in the reprogramming of host defense and metabolism are gradually emerging. Given the plethora of secreted proteins encoded in the genomes of phytopathogenic fungi, only a few fungal effector targets have been identified to date, with less than a handful shown to impair host metabolic functions directly (Djamei et al., 2011; Hemetsberger et al., 2012; Tanaka et al., 2014). Although eight AVR genes of M. oryzae have been isolated over the last several decades (Zhang et al., 2015), their functions as virulence factors have not been functionally addressed, except for evidences found in AVR-Pizt and AVR-Pii (Fujisaki et al., 2015; Park et al., 2012).
The Y2H, BiFC, Co-IP and GST pull-down data of AVR-Pii and Os-NADP-ME2-3 strongly support the notion that AVR-Pii interacts with Os-NADP-ME2-3 in the rice cytoplasm (Figs. 1 and 2; Supplementary Fig. S2). This is in agreement with the data from Parker et al. (2009), whereby Os-NADP-MEs activities are localized at the penetration site following M. oryzae inoculation. These protein–protein interactions at the infection site could reprogram host metabolism by suppressing localized Os-NADP-ME2-3 activities and significantly decrease NADPH production.
A decrease in ROS accumulation occurred in susceptible cultivar at 24 h post-inoculation (hpi), but no change in ROS concentration was observed in resistant cultivar following M. oryzae inoculation in Brachypodium distachyon (Parker et al., 2009). This characteristic ROS suppression in a susceptible cultivar could be explained by our results, whereby M. oryzae secretes AVR-Pii during BIC formation to inhibit Os-NADP-ME2-3 activity in infected cells and a few neighboring cells (Fig. 4B). This protein–protein interaction-triggered inhibition of Os-NADP-ME2-3 could decrease cytoplasmic NADPH production and the consequent electron transfer that is an indispensable part of ROS metabolism. This mechanism also explains the active suppression of the typical second wave of ROS production by virulent pathogens (Shaw, 2003). In fact, ROS production in the ΔOs-nadp-me2-3 mutant by PAMP elicitor, chitin was significantly decreased (Fig. 5C). This indicates that Os-NADP-ME2-3 is directly associated with ROS burst and is also important in PTI, as suggested in Arabidopsis NADP-ME2 (Voll et al., 2012). Thus, Os-NADP-ME2-3-mediated ROS burst following PAMP reception may play an essential role in the exclusion of pathogens and subsequent activation of plant defense responses (Kadota et al., 2014; Levine et al., 1994; Torres et al., 2006; Voll et al., 2012). According to Chi et al. (2009), in vitro H2O2 inhibits M. oryzae growth at low concentrations, suggesting that M. oryzae IH growth is significantly suppressed when H2O2 concentrations reach inhibitory levels in the EIHMx under normal ROS burst conditions. Another important point is that reduced ROS is only one of many potential explanations for the effector virulence functions. Therefore, the data to date do not clearly identify which is the most important virulence functions for infection. NADP-malic enzymes are also found within a wide range of eukaryotic organisms, including fungi themselves. Based on the recent reported data on activation of ROS after PAMP perception (Li et al., 2014) and by ETI through MAPKs signaling pathway (Adachi et al., 2015), it seems like that they are self controlled under normal condition but there are various ways to regulate ROS production from inactive to active conditions. Thus, self control of NADP-malic enzyme is one of the possible mechanisms to maintain optimal ROS production in eukaryotic organisms.
AVR effectors have dual activities: they suppress PTI thereby enhancing pathogenesis in host cells in the absence of its cognate R genes, and they trigger rapid HR in the presence of R proteins by ETI (Göhre and Robatzek, 2008). OsExo70-F2 which is also identified in our study as AVR-Pii interacting protein has been suggested as an additional factor in ETI immunity by interacting with AVR-Pii (Fujisaki et al., 2015).We observed distinct AVR-Pii expression as well as typical rice blast symptoms (Figs. 3B and 4B), as expected from its virulence function in susceptible rice cultivar NB (Fujisaki et al., 2015). However, in the ΔOs-nadp-me2-3 mutant, despite no visible AVR-Pii expression, there were significantly increased viable IHs (Figs. 3C and 4B). We do not know why AVR effector expression was undetected in the ΔOs-nadp-me2-3 mutant, but the other AVR effector, PWL2, showed the same undetectable expression pattern (Supplementary Fig. S9B). This suggests that host ROS itself together with ROS-related in planta condition during infection may influence the sophisticated AVR effector regulatory mechanism as signal molecules. Suppression of ROS production in ΔOs-nadp-me2-3 may negatively regulate AVR effector expression via a secretion mechanism that is largely unknown. Because we only examined a limited sample, more experimental data are required to address whether this ATRI mechanism is employed as a common strategy of AVR effectors. Regarding other virulence functions of AVR-Pii, ectopic expression of AVR-Pii in susceptible backgrounds did not affect the pathogenicity of a compatible isolate of M. oryzae (Fujisaki et al., 2015). Therefore, it is believed that the contribution of AVR-Pii to pathogen virulence cannot be readily established under the current laboratory conditions.
Our data agree with previous OsRac1-mediated ROS production and cell death mechanisms (Kawasaki et al., 1999), which involve a positive regulator of resistance to fungal and bacterial pathogens (Ono et al., 2001). Rice OsRac1 regulates ROS production by directly interacting with the N-terminus of NADPH-oxidase (Wong et al., 2004) and forming a putative immune complex at the plasma membrane with RACK1A, RAR1, SGT1, HSP90, and HSP70 (Nakashima et al., 2008). The interaction of tobacco NADP-ME with HSP70 (Lara et al., 2005) suggests that Os-NADP-ME2 interacts with Nakashima’s immune complexes (Nakashima et al., 2008), which were recently described as a defensome (Akamatsu et al., 2013). This indicates that Os-NADP-ME2 can directly regulate ROS production in the defensome complex to control innate immunity. Recent data support that massive ROS bursts could inactivate infecting M. oryzae in the EIHMx (Parker et al., 2009), which is highly similar to the animal phagosome that generates the superoxide anion by consuming NADPH through plasma membrane-bound NADPH-oxidase enzyme complexes (Lambeth, 2004). ROS could be produced continuously with NADP-ME-assisted NADPH to plasma membrane-bound NADPH-oxidase enzymes (Parker et al., 2009). Thus, the depletion of cytoplasmic NADPH could negatively influence the PAMP-triggered ROS burst, which has been detected in the invaded cell as well as neighboring cells (Parker et al., 2009; Voll et al., 2012). Indeed, our PAMP-triggered ROS assay data (Fig. 5C) strongly support the fact that AVR-Pii-triggered Os-NADP-ME2-3 inhibition attenuates ROS burst and helps M. oryzae establish compatibility in susceptible rice cultivar (Fig. 3). Meanwhile, we couldn’t ignore the diverse virulence functions of AVR effectors in collaboration with their interacting proteins in immune responses. It is also known that single effector can interacts with multiple targets and each interaction may have individual contribution for virulence. For example, in our study we showed that AVR-Pii attenuates host ROS burst by targeting and inhibiting OsNADP-ME2-3 which is indispensable in ROS metabolism. Similarly it has been suggested that, AVR-Pii also targets OsEXO70-F3 (Fujisaki et al., 2015). The relationship between the AVR-Pii-triggered ROS inhibition by targeting OsNADP-ME2-3 and the role of OsExo70 as a helper in Pii/AVR-Pii interactions is another interesting area for further study. It will be interesting to investigate whether AVR-Pii/OsExo70-mediated virulence function and AVR-Pii/OsNADP-ME2-3-mediated virulence function is dependent or independent of each other. Comparative analyses of the virulence function of AVR-Pii/OsNADP-ME2-3 pair and AVR-Pii/OsExo70 pair will provide new insight into differentiating downstream responses between PTI and ETI respectively. Due to the lack of enough virulence functional resources on AVR effectors, these two individual targets (OsNADP-ME2-3 and OsExo70) of AVR-Pii will definitely increase our understanding of virulence functions of effectors more effectively.
Based on our results, we generated a model of AVR-Pii and Os-NADP-ME interactions near the BIC after infection. In our model, M. oryzae INA168 secretes AVR-Pii through the BIC to disrupt ROS burst by suppressing NADPH production in susceptible rice cultivar (Fig. 7). The AVR-Pii-Os-NADP-ME2-3 pair provides key evidence on how AVR effectors reprogram host metabolism to establish compatibility and why the ROS issue has continued to be treated as a main defense mechanism in the innate immunity of most plants.
Acknowledgments
This work was supported by a grant from Republic of Korea and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (Grant No. 2013R1A1A 2009269). We thank Drs. An and J.S. Jeon for providing the T-DNA insertion mutant Os-nadp-me with HY background.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Able A.J. Role of reactive oxygen species in the response of barley to necrotrophic pathogens. Protoplasma. 2003;221:137–143. doi: 10.1007/s00709-002-0064-1. [DOI] [PubMed] [Google Scholar]
- Adachi H., Nakano T., Miyagawa N., Ishihama N., Yoshioka M., Katou Y., Yaeno T., Shirasu K., Yoshioka H. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell. 2015;27:2645–2663. doi: 10.1105/tpc.15.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu A., Wong H.L., Fujiwara M., Okuda J., Nishide K., Uno K., Imai K., Umemura K., Kawasaki T., Kawano Y., Shimamoto K. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitininduced rice immunity. Cell Host Microbe. 2013;13:465–476. doi: 10.1016/j.chom.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Cesari S., Thilliez G., Ribot C., Chalvon V., Michel C., Jauneau A., Rivas S., Alaux L., Kanzaki H., Okuyama Y., et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell. 2013;25:1463–1481. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S., Bernoux M., Moncuquet P., Kroj T., Dodds P.N. A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 2014a;5:606. doi: 10.3389/fpls.2014.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S., Kanzaki H., Fujiwara T., Bernoux M., Chalvon V., Kawano Y., Shimamoto K., Dodds P., Terauchi R., Kroj T. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014b;33:1941–1959. doi: 10.15252/embj.201487923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Yang J., Wu N., Zhang F. Four rice genes encoding NADP malic enzyme exhibit distinct expression profiles. Biosci. Biotechnol. Biochem. 2004;68:1865–1874. doi: 10.1271/bbb.68.1865. [DOI] [PubMed] [Google Scholar]
- Chi M.H., Park S.Y., Kim S., Lee Y.H. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 2009;5:e1000401. doi: 10.1371/journal.ppat.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Horvath D.M., Staskawicz B.J. Pivoting the plant imuune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 2011;35:1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- Detarsio E., Andreo C.S., Drincovich M.F. Basic residues play key roles in catalysis and NADP+-specificity in maize (Zea mays L.) photosynthetic NADP+-dependent malic enzyme. Biochem. J. 2004;382:1025–1030. doi: 10.1042/BJ20040594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A., Schipper K., Rabe F., Ghosh A., Vincon V., Kahnt J., Osorio S., Tohge T., Fernie A.R., Feussner I., et al. Metabolic priming by a secreted fungal effector. Nature. 2011;478:395–398. doi: 10.1038/nature10454. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., van der Linde K., Assmann D., Schwammbach D., Hof A., Mohanty A., Jackson D., Kahmann R. Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 2009;5:e1000290. doi: 10.1371/journal.ppat.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke N. Involvement of superoxide anion genertaion in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 1983;23:345–357. [Google Scholar]
- Edwards G.E., Andreo C.S. NADP-malic enzyme from plants. Phytochemistry. 1992;31:1845–1857. doi: 10.1016/0031-9422(92)80322-6. [DOI] [PubMed] [Google Scholar]
- Fisher M.C., Henk D.A., Briggs C.J., Brownstein J.S., Madoff L.C., McCraw S.L., Gurr S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971;9:275–296. [Google Scholar]
- Fujisaki K., Abe Y., Ito A., Saitoh H., Yoshida K., Kanzaki H., Kanzaki E., Utsushi H., Yamashita T., Kamoun S., et al. Rice Exo70 interacts with a fungal effector, AVR-Pii and is required for AVR-Pii-triggered immunity. Plant J. 2015;83:875–887. doi: 10.1111/tpj.12934. [DOI] [PubMed] [Google Scholar]
- Gabriel D.W., Rolfe B.G. Working models of specific recognition in plant-mirocbe interactions. Annu. Rev. Phytopathol. 1990;28:365–391. [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.R., Hansch R. New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant. 2009;2:1051–1058. doi: 10.1093/mp/ssp040. [DOI] [PubMed] [Google Scholar]
- Giraldo M.C., Valent B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013;11:800–814. doi: 10.1038/nrmicro3119. [DOI] [PubMed] [Google Scholar]
- Giraldo M.C., Dagdas Y.F., Gupta Y.K., Mentlak T.A., Yi M., Martinez-Rocha A.L., Saitoh H., Terauchi R., Talbot N.J., Valent B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 2013;4:1996. doi: 10.1038/ncomms2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V., Robatzek S. Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 2008;46:189–215. doi: 10.1146/annurev.phyto.46.120407.110050. [DOI] [PubMed] [Google Scholar]
- Grant J.J., Loake G.J. Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 2000;124:21–29. doi: 10.1104/pp.124.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg J.T., Yao N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- Hemetsberger C., Herrberger C., Zechmann B., Hillmer M., Doehlemann G. The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 2012;8:e1002684. doi: 10.1371/journal.ppat.1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Lee S., Yang K., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeon J., Goh J., Yoo S., Chi M.H., Choi J., Rho H.S., Park J., Han S.S., Kim B.R., Park S.Y., et al. A putative MAP kinase kinase kinase, MCK1, is requried for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant Microbe Interact. 2008;21:525–534. doi: 10.1094/MPMI-21-5-0525. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015;56:1472–1480. doi: 10.1093/pcp/pcv063. [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007;19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T., Henmi K., Ono E., Hatakeyama S., Iwano M., Satoh H., Shimamoto K. The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.Y., Czymmek K., Kang S., Valent B. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell. 2010;22:1388–1403. doi: 10.1105/tpc.109.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.A., Cho K., Singh R., Jung Y.H., Jeong S.H., Kim S.H., Lee J., Cho Y.S., Agrawal G.K., Rakwal R., et al. Rice OsACDR1 (Oryzae sativa accelerated cell death and resistance 1) is a potential positive regulator of fungal disease resistance. Mol. Cells. 2009;28:431–439. doi: 10.1007/s10059-009-0161-5. [DOI] [PubMed] [Google Scholar]
- Lambeth J.D. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- Lara M.V., Drincovich M.F., Muller G.L., Maurino V.G., Andreo C.S. NADP-malic enzyme and Hsp70: copurification of both proteins and modification of NADP-malic enzyme properties by association with Hsp70. Plant Cell Physiol. 2005;46:997–1006. doi: 10.1093/pcp/pci108. [DOI] [PubMed] [Google Scholar]
- Levine A., Tenhaken R., Dixon R., Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15:329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Liu J., Wang X., Mitchell T., Hu Y., Liu X., Dai L., Wang G.L. Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol. Plant Pathol. 2010;11:419–427. doi: 10.1111/j.1364-3703.2009.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciel J.L., Ceresini P.C., Castroagudin V.L., Zala M., Kema G.H., McDonald B.A. Population structure and pathotype diversity of the wheat blast pathogen Magnaporthe oryzae 25 years after its emergence in Brazil. Phytopathology. 2014;104:95–107. doi: 10.1094/PHYTO-11-12-0294-R. [DOI] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Maqbool A., Saitoh H., Franceschetti M., Stevenson C.E., Uemura A., Kanzaki H., Kamoun S., Terauchi R., Banfield M.J. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife. 2015;4 doi: 10.7554/eLife.08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D., Dunand C., Puppo A., Pauly N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012;17:9–15. doi: 10.1016/j.tplants.2011.10.001. [DOI] [PubMed] [Google Scholar]
- McHale L., Tan X., Koehl P., Michelmore R.W. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera G., Giraldo M.C., Khang C.H., Coughlan S., Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as Biotrophy-associated secreted proteins in rice blast disease. Plant Cell. 2009;21:1273–1290. doi: 10.1105/tpc.107.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Chen L., Thao N.P., Fujiwara M., Wong H.L., Kuwano M., Umemura K., Shirasu K., Kawasaki T., Shimamoto K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell. 2008;20:2265–2279. doi: 10.1105/tpc.107.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E., Wong H.L., Kawasaki T., Hasegawa M., Kodama O., Shimamoto K. Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA. 2001;98:759–764. doi: 10.1073/pnas.021273498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.H., Chen S., Shirsekar G., Zhou B., Khang C.H., Songkumarn P., Afzal A.J., Ning Y., Wang R., Bellizzi M., et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogenassociated molecular pattern-triggered immunity in rice. Plant Cell. 2012;24:4748–4762. doi: 10.1105/tpc.112.105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Jeong M.H., Wang H.Y., Kim J.A., Yu N.H., Kim S., Cheong Y.H., Kang S., Lee Y.H., Hur J.S. Agrobacterium tumefaciens-mediated transformation of the lichen fungus, Umbilicaria muehlenbergii. PLoS One. 2013;8:e83896. doi: 10.1371/journal.pone.0083896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D., Beckmann M., Zubair H., Enot D.P., Caracuel-Rios Z., Overy D.P., Snowdon S., Talbot N.J., Draper J. Metabolomic analysis reveals a common pattern of metabolic reprogramming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009;59:723–737. doi: 10.1111/j.1365-313X.2009.03912.x. [DOI] [PubMed] [Google Scholar]
- Piedras P., Hammond-Kosack K.E., Jones J.D.G. Rapid, Cf-9-and Avr9-dependent production of active oxygen species in tobacco suspension cultures. Mol. Plant Microbe Interact. 1998;11:1155–1166. [Google Scholar]
- Pogány M., von Rad U., Grün S., Dongó A., Pintye A., Simoneau P., Bahnweg G., Kiss L., Barna B., Durner J. Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 2009;151:1459–1475. doi: 10.1104/pp.109.141994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Sharma S., Hirabuchi A., Yoshida K., Fujisaki K., Ito A., Uemura A., Terauchi R., Kamoun S., Sohn K.H., et al. Deployment of the Burkholderia glumae type III secretion system as an efficient tool for translocating pathogen effectors to monocot cells. Plant J. 2013;74:701–712. doi: 10.1111/tpj.12148. [DOI] [PubMed] [Google Scholar]
- Shaw S.L. Nod factor inhibition of reactive oxygen efflux in a host legume. Plant Physiol. 2003;132:2196–2204. doi: 10.1104/pp.103.021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Lee M.O., Lee J.E., Choi J., Park J.H., Kim E.H., Yoo R.H., Cho J.I., Jeon J.S., Rakwal R., et al. Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 2012;160:477–487. doi: 10.1104/pp.112.200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Dangol S., Jwa N.S. Yeast two-hybrid system for dissecting the rice MAPK interactome. Methods Mol. Biol. 2014a;1171:195–216. doi: 10.1007/978-1-4939-0922-3_16. [DOI] [PubMed] [Google Scholar]
- Singh R., Lee J.E., Dangol S., Choi J., Yoo R.H., Moon J.S., Shim J.K., Rakwal R., Agrawal G.K., Jwa N.S. Protein interactome analysis of 12 mitogen-activated protein kinase kinase kinase in rice using a yeast two-hybrid system. Proteomics. 2014b;14:105–115. doi: 10.1002/pmic.201300125. [DOI] [PubMed] [Google Scholar]
- Stael S., Kmiecik P., Willems P., Van Der Kelen K., Coll N.S., Teige M., Van Breusegem F. Plant innate immunity--sunny side up? Trends Plant Sci. 2015;20:3–11. doi: 10.1016/j.tplants.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Brefort T., Neidig N., Djamei A., Kahnt J., Vermerris W., Koenig S., Feussner K., Feussner I., Kahmann R. A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. Elife. 2014;3:e01355. doi: 10.7554/eLife.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005;37:1130–1134. doi: 10.1038/ng1639. [DOI] [PubMed] [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;141:373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B., Khang C.H. Recent advances in rice blast effector research. Curr. Opin. Plant. Biol. 2010;13:434–441. doi: 10.1016/j.pbi.2010.04.012. [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A., Kamoun S. From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll L.M., Zell M.B., Engelsdorf T., Saur A., Wheeler M.G., Drincovich M.F., Weber A.P., Maurino V.G. Loss of cytosolic NADP-malic enzyme 2 in Arabidopsis thaliana is associated with enhanced susceptibility to Colletotrichum higginsianum. New Phytol. 2012;195:189–202. doi: 10.1111/j.1469-8137.2012.04129.x. [DOI] [PubMed] [Google Scholar]
- Wang W., Wen Y., Berkey R., Xiao S. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell. 2009;21:2898–2913. doi: 10.1105/tpc.109.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Ou B., Li J., Zhao Y., Guo D., Zhu Y., Chen Z., Gu H., Li C., Qin G., et al. Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance. PLoS One. 2013;8:e59720. doi: 10.1371/journal.pone.0059720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.C., Tronconi M.A., Drincovich M.F., Andreo C.S., Flügge U.I., Maurino V.G. A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol. 2005;139:39–51. doi: 10.1104/pp.105.065953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.J., Sohn K.H., Wan L., Bernoux M., Sarris P.F., Segonzac C., Ve T., Ma Y., Saucet S.B., Ericsson D.J., et al. Structural basis for assembly and function of heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- Wong H.L., Sakamoto T., Kawasaki T., Umemura K., Shimamoto K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004;135:1447–1456. doi: 10.1104/pp.103.036384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Saitoh H., Fujisawa S., Kanzaki H., Matsumura H., Yoshida K., Tosa Y., Chuma I., Takano Y., Win J., et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell. 2009;21:1573–1591. doi: 10.1105/tpc.109.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang L., Wu W., He L., Yang X., Pan Q. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene. Pib. Sci. Rep. 2015;5:11642. doi: 10.1038/srep11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


