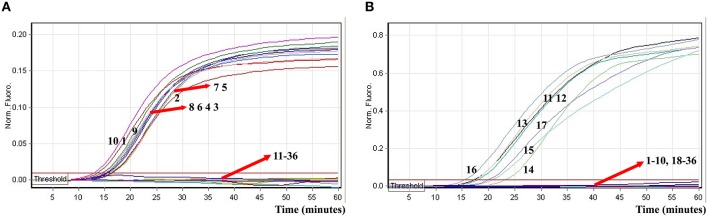
Figure 9.
The analytical specificity of multiplex ET-MCDA detection for different strains. The multiplex ET-MCDA amplifications were conducted using different genomic DNA templates and were monitored by means of real-time format. (A,B) were simultaneously generated from HEX and Cy5 channels. Signals 1-10, Shigella flexneri strains of serovar 1d (ICDC-NPS001), 4a (ICDC-NPS002), 5a (ICDC-NPS003), 2b (ICDC-NPS004), 1b (ICDC-NPS005), 3a (ICDC-NPS006), 3b (ICDC-NPS008), Shigella boydii, Shigella sonneri, and Shigella dysenteriae; signals 11–15, Salmonella Choleraesuis (ICDC-NPSa001), Salmonella Dublin (ICDC-NPSa002), Salmonella Enteritidis (ICDC-NPSa003), Salmonella Typhimurium (ICDC-NPSa004), Salmonella Weltevreden (ICDC-NPSa005); signals 16 and 17, two Salmonella strains of unidentified serotype; signals 18-35, Listeria monocytogenes stains of serovar 1/2a (EGD-e), 4c (19116), Enteroinvasive E. coli, Enteropathogenic E. coli, Enterotoxigenic E. coli, Enteroaggregative E. coli, Enterohemorrhagic E. coli, Plesiomonas shigelloides, Yersinia enterocolitica, Bntorobater sakazakii, Enterobacter cloacae, Enterococcus faecalis, Vibrio parahaemolyticus, Vibrio vulnificus, Bacillus cereus, Staphylococcus aureus, Campylobacter jejuni, and Pseudomonas aeruginosa, signal 36, negative control.