Fig. 1.
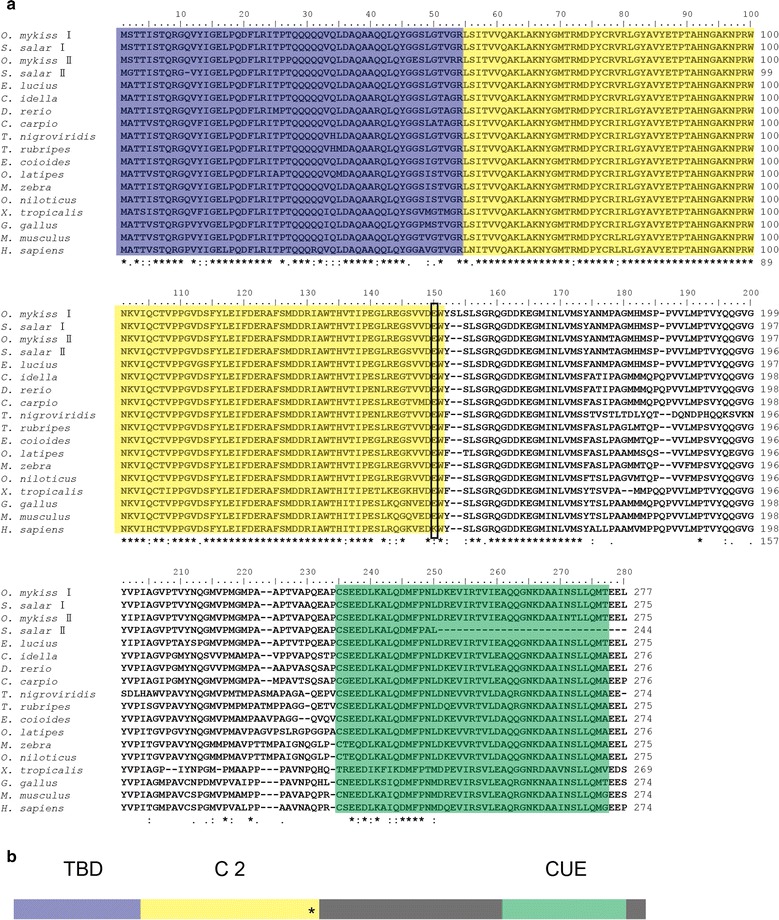
Multiple alignment and schematic representation of Tollip. a Amino acid sequence alignment and functional domains of Tollip. The sequences were aligned by ClustalX 2.0. The identical amino acid residue is indicated in the asterisk (*); the predicted domains from SMART Server software have been indicated by colored boxes: the blue denotes the TBD (Tom1-binding domain), the yellow shows the C2 (conserved core 2) domain, the green indicates the CUE (coupling of ubiquitin to endoplasmic reticulum degradation) domain. The black box points the amino acid at position 150. The GenBank accession numbers of these sequences are listed in Additional file 1: Table S1. b Schematic representation of the structural domains of Tollip. The predicted domains from SMART Server software are indicated by colored boxes as shown in Fig. 1a, while other regions are shaded in grey color. The asterisk (*) represent the amino acid at the position 150
