Fig. 6.
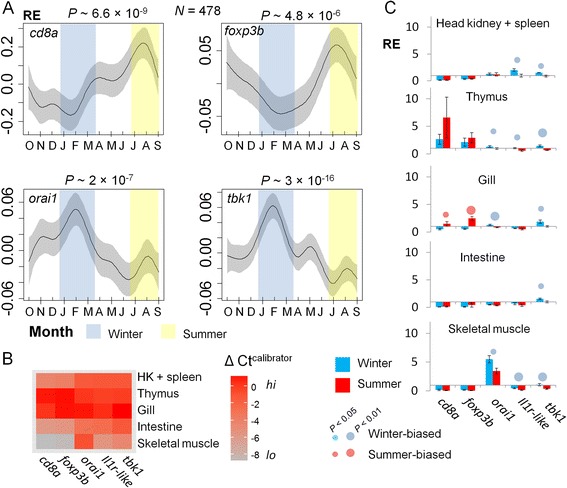
Corroborative whole-fish and tissue-specific Q-PCR gene expression measurements. a Temporal variation from October 2013 to September 2014 in whole-fish gene expression for winter-summer interface (key) genes from Network 1, n = 478. Relative expression (RE) (normalized to endogenous control genes and indexed to a calibrator sample) is indicated on the y-axis. Plots show thin-plate-spline smoothers for time fitted in a generalized additive model (GAM) with fixed effects for habitat, sex and length; shaded areas represent 95 % confidence regions. Samples were derived from FRN, RHD and artificial outdoors habitats stocked from FRN. The other key gene examined, il1r-like, also demonstrated significant seasonal variation (P = 0.0019) with peak expression in December (not shown), if log10 transformed. b Tissue-specific expression of key genes at STO (summer, n = 5; winter n = 10). Heat map showing relative gene expression across tissues; significant differences occurred for all genes (P <0.001). c Tissue-specific seasonal variation in key genes at STO. Mean relative expression (RE) ± 1 SE is shown on the y-axis. P values (c) relate to directional (1-tailed) t-tests of seasonal shifts in the same direction as the whole-fish RNAseq study; 13/25 of these tests were significant but, in comparison (post-hoc), 0/25 of 1-tailed tests in the opposite direction were significant. The calibrator sample for tissue-specific analyses was pooled whole-fish RNA from 20 individuals in September. The STO samples showed no significant difference in fish length between winter and summer and were balanced for sex ratio (see Additional file 8: Table S6)
