Fig. 3.
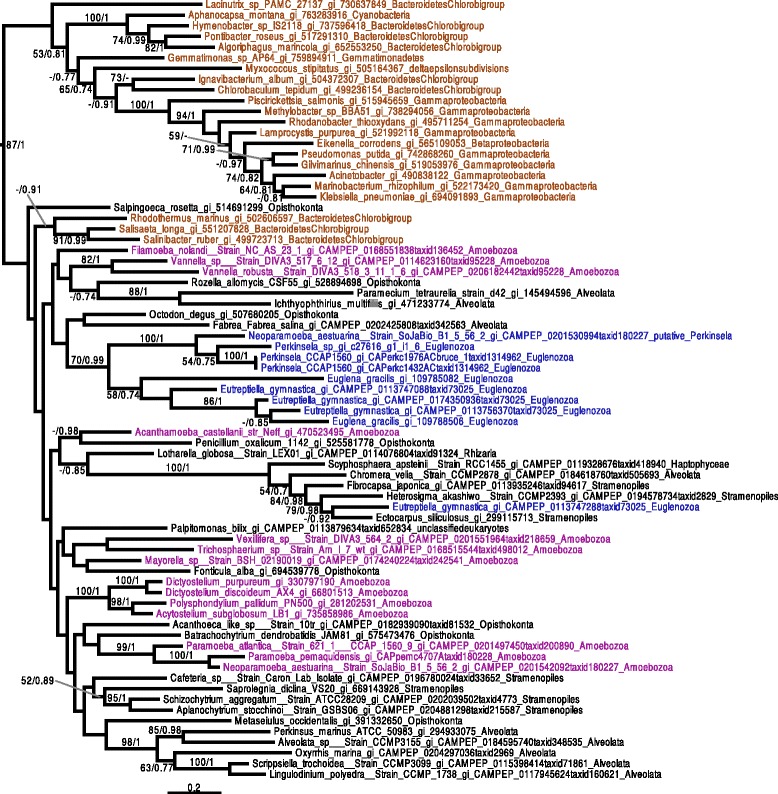
UROD phylogenetic tree. The tree shown is the consensus tree obtained with Phylobayes 4.1 with ML boostrap values (left) and Bayesian posterior probabilities (right) mapped onto the nodes. Bootstrap values >50 % are shown, while only posterior probabilities >0.6 are shown. The tree is rooted with the proteobacterial sequences, as in Kořený and Oborník [67]. Sequences are colored according to their taxonomic affiliation: Amoebozoa are in purple, Euglenozoa are in blue, other Eukaryota are in black, and Bacteria are brown. A trio of sequences from the euglenozoan Eutreptiella gymnastica, as well as two Euglena gracilis sequences, group with Prokinetoplastina with a bootstrap value of 70 % and a posterior probability of 0.99. Another E. gymnastica sequence branches elsewhere in the tree. The scale bar shows the inferred number of amino acid substitutions per site
