Fig. 7.
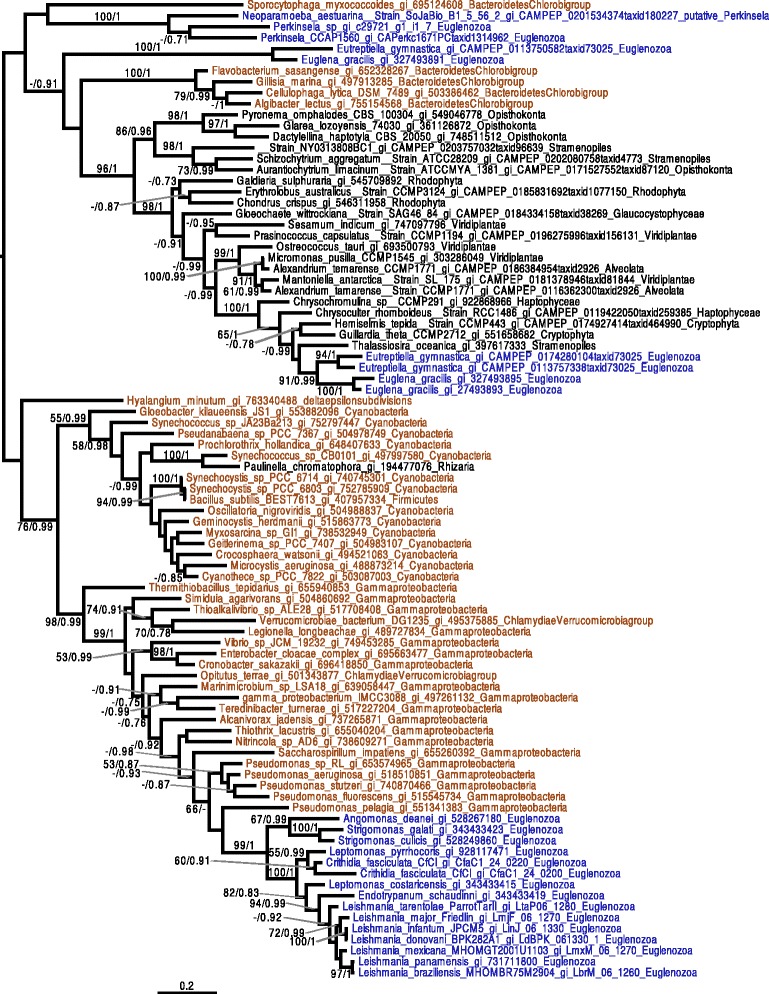
CPOX/HemF phylogenetic tree. The tree shown is the consensus tree obtained with Phylobayes 4.1 with ML boostrap values (left) and Bayesian posterior probabilities (right) mapped onto the nodes. Bootstrap values >50 % are shown, while only posterior probabilities >0.6 are shown. Groups are colored depending on their taxonomic group: Euglenozoa are in blue, other Eukaryotes are in black while Bacteria are colored brown. The Perkinsela spp. sequences group near the other eukaryotic sequences, albeit intermingled with bacterial sequences. The Leishmaniinae/Strigomonadinae sequences are nested within Gammaproteobacteria. The scale bar shows the inferred number of amino acid substitutions per site
