Fig. 8.
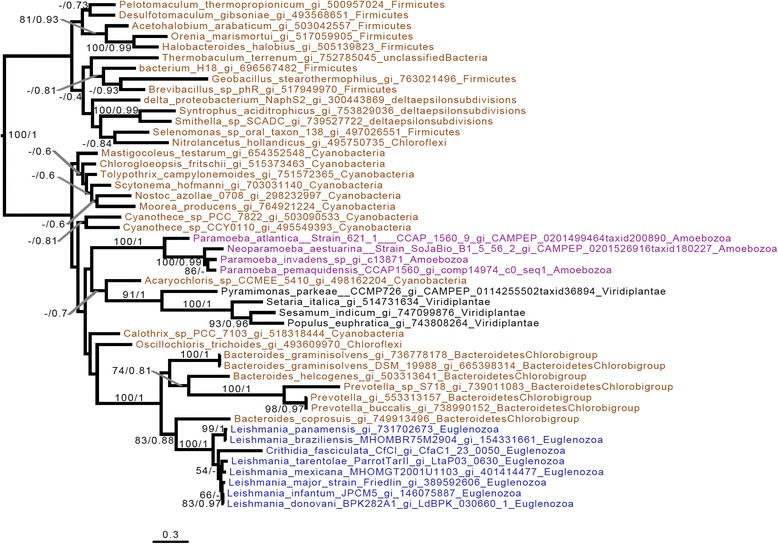
UROS phylogenetic tree. The tree shown is the consensus tree obtained with Phylobayes 4.1 with ML bootstrap values (left) and Bayesian posterior probabilities (right) mapped onto the nodes. Bootstrap values >50 % are shown, while only posterior probabilities >0.6 are shown. The tree is rooted with a distant bacterial group composed of Firmicutes/Proteobacteria/Chloroflexi. Taxonomic labels are as follows: Bacteria are brown, Euglenozoa are in blue, Amoebozoa are in purple, and other eukaryotes are in black. The sequences of the kinetoplastid genus Leishmania branch with a group of Bacteroidetes, which belong itself to a bigger group composed of cyanobacteria, Viridiplantae, and the Paramoeba spp. Perkinsela spp. sequences are not included, as no UROS genes were detected. The scale bar shows the inferred number of amino acid substitutions per site
