Figure 4. Protein Occupancy and Secondary Structure Landscapes at Alternative Splicing and Polyadenylation Sites.
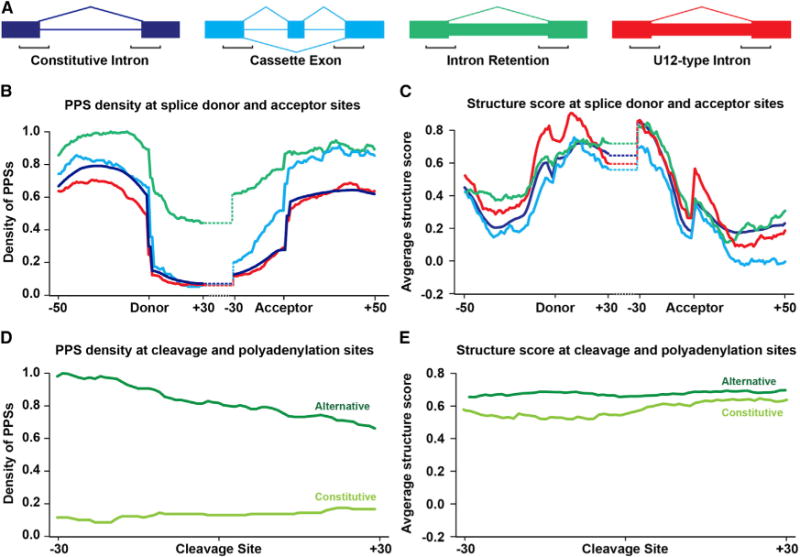
(A) Diagram of constitutive introns (blue), cassette exons (turquoise), intron retention events (green), and U12-type introns (red). Large boxes represent exons, lines represent constitutive introns, and small boxes represent alternatively spliced sequences, with the black brackets indicating the regions graphed in (B) and (C) for reference.
(B) PPS density profiles for constitutive and alternative splicing events in Arabidopsis. Average PPS density at each position −50 to +30 nt at the donor splice site, and −30 to +50 nt at the acceptor splice site. Line colors correspond to examples shown in (A).
(C) Structure score profiles for constitutive and alternative splicing events in Arabidopsis covering the same regions as (B). Line colors correspond to examples shown in (A).
(D) PPS density profiles for constitutive and alternative poly(A) sites of nuclear mRNAs. Average PPS density at each position ± 30 nt from constitutive (light-green line) and alternative (dark-green line) cleavage and polyadenylation sites.
(E) Average structure score profiles for constitutive and APA sites covering the same regions as (D). See also Figure S6.
