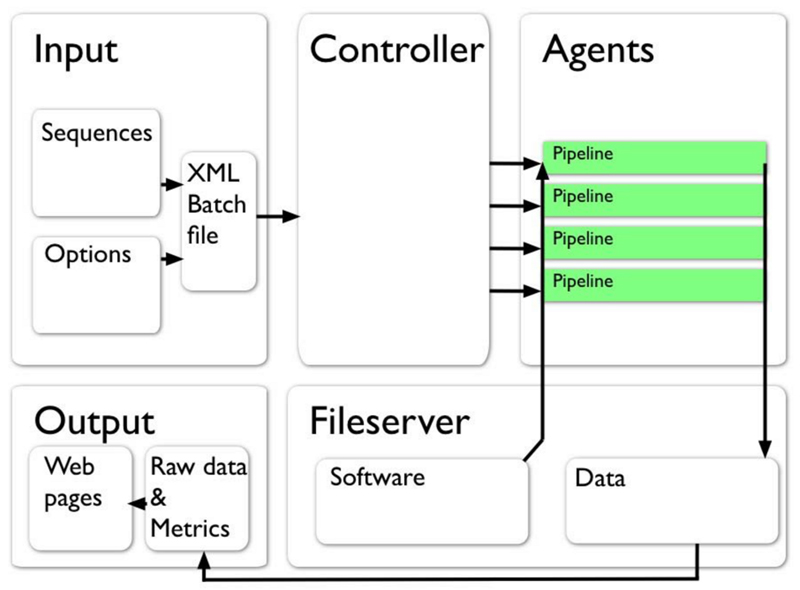
Figure 3.
Schematic of the ensemble simulation workflow (arrows represent the flow and transformation of information at distinct stages of the ensemble). To generate an ensemble of Sidekick jobs a collection of helix amino acid sequences and simulation options (e.g. number of jobs, choice of forcefield, etc.) are used to generate an XML input file for submission to the controller, a machine which coordinates jobs across the grid. From this file the controller starts jobs on all available “agents”, which run individual pipelines, reading required software from a centralised fileserver and depositing output files onto the same fileserver. Files may be written locally during the simulation and copied at the end of the job, or written as the job is running. Individual simulations are analysed within each pipeline to generate metrics to describe the orientation of the helix over the course of the simulation. Both raw data (trajectories and other outputs) and descriptive metrics (e.g. the position and orientation of the helix in the membrane) can be accessed on the fileserver or presented through a web interface.