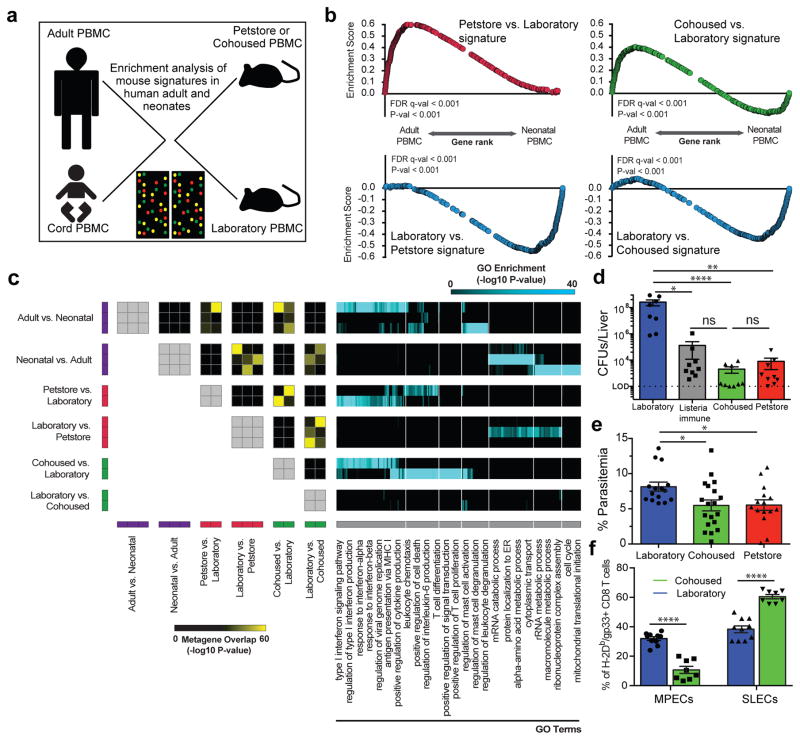
Figure 4. Microbial experience matures mouse immune transcriptome from neonatal to adult human and impacts immune system function.
a) Experimental design and b) GSEA plots showing the enrichment of gene signatures among the indicated mouse group comparisons relative to human adult vs. neonatal comparison. Signatures consist of top 400 significantly differentially expressed genes of laboratory (n=8), cohoused (n=7) and petstore mice (n=8). c) Pairwise overlaps of metagenes identified through leading-edge metagene analysis and corresponding GO terms. d) Bacterial load in the liver 3 days post-challenge with 8.5 × 104 CFU of Listeria monocytogenes (LM) among indicated groups (n=9 for all except laboratory mice where n=8). e) Parasitic load in peripheral blood 5 days post Plasmodium berghei ANKA parasitized RBC challenge among the indicated groups of laboratory (n=15), cohoused (n=19), and petstore mice (n=15). f) 28 days after LCMV Armstrong infection, the proportion of H-2Db/gp33-specific CD8 T cell MPECs (KLRG1-, CD127+) and SLECs (KLRG1+, CD127−) in PBMC of cohoused (n=8) and laboratory (n=9). Data are cumulative of 2 independent experiments. d–f) Data points represent individual mice. Significance was determined using Kruskal-wallis (ANOVA) test (in d), one-way ANOVA test (in e) and unpaired two-sided t-test (in f) * p<0.05, ** p<0.01, **** p<0.0001. Bars indicate mean ± S.E.M.