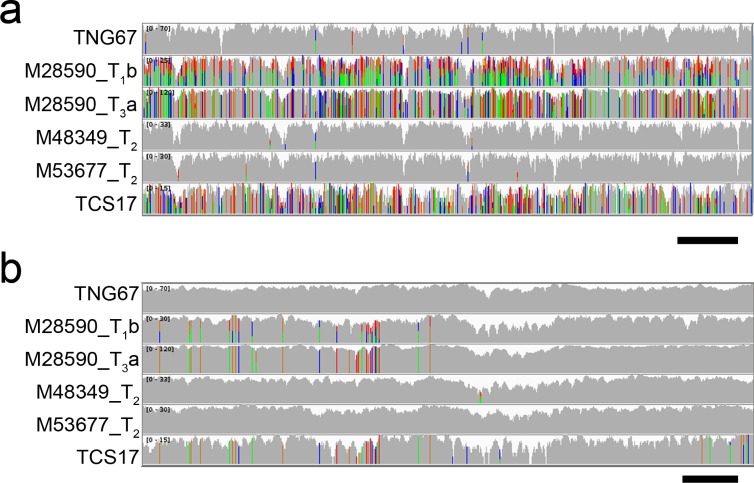
Fig 2. Introgression in M0028590 offspring.
The Integrative Genomics Viewer (IGV) view of aligned reads. Panel a. A 100-kb region in chromosome 2 (35,200,001–35,300,000). The paired read sequencing was performed on an Illumina platform and the alignments were toward the Nipponbare IRGSP 1.0. The x-axis is the thickness of the read (shown in log scale), with the gray bar for matched reads (compared to Nipponbare); the mismatched reads for A are in green, G in orange, C in blue, and T in red. Rows a to f, Tainung 67 (TNG67), M0028590T1b, M0028590T3a, a T2 plant of M0048349, a T2 plant of M0053677, Taichungsen 17 (TCS17). Scale bar: 10 kb. Panel b. IGV view of aligned reads of a 10-kb region of chromosome 2 (35,420,601–35,431,600). Scale bar: 1 kb.