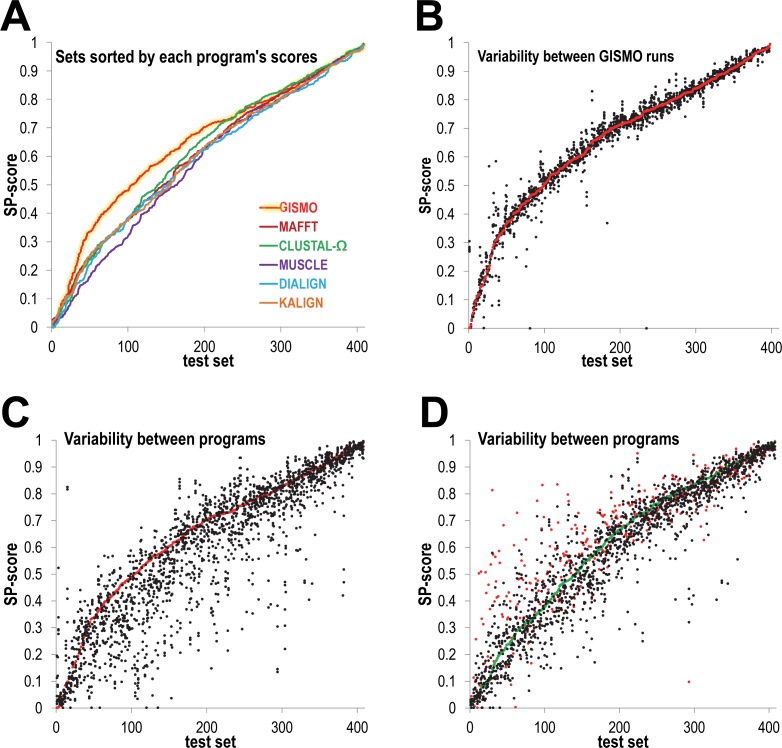
Fig 3. Variability in SP-scores among six GISMO runs and among the six programs GISMO, MAFFT, CLUSTAL-Ω, MUSCLE, Dialign and Kalign.
SP-scores are based upon the CDD MSAs as benchmarks and vary from 0 (no correctly aligned sequence pairs) to 1 (all pairs aligned correctly). A. The sorted SP-scores for a single GISMO run (red line with yellow back-glow) compared with the sorted scores for the five other programs. B. Run-to-run variability in SP-scores over six GISMO runs. Test set data points are sorted along the x-axis by the SP-score obtained for each set on the first run (red data points) of six. C. SP-scores for the six programs analyzed, sorted by the GISMO score on each test set. GISMO SP-scores (for a single run) are shown in red. Each red data point and the five black data points (one point for each program) plotted in the same column correspond to the same test set. D. SP-scores for the six programs, sorted by the CLUSTAL-Ω score on each test set. Data points for GISMO and for CLUSTAL-Ω are shown in red and green, respectively.