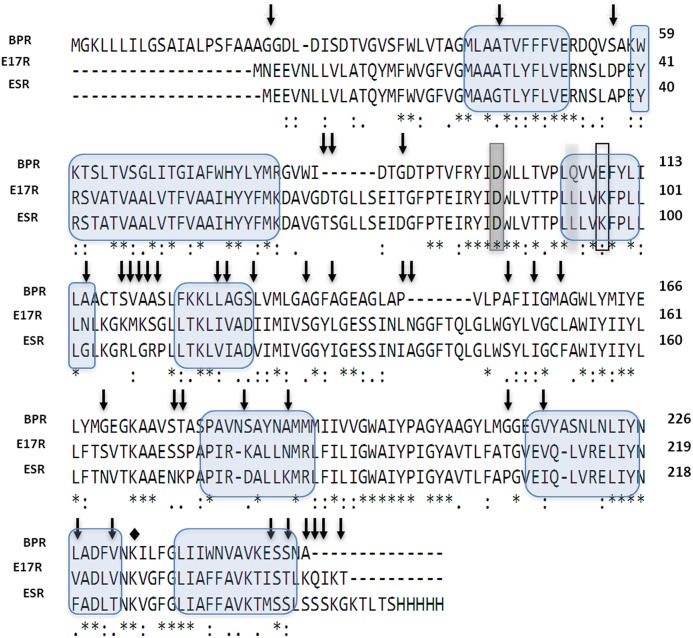
Fig 2. Multiple protein alignment of PR from Exiguobacterium sp. S17 (E17R), green-light absorbing proteorhodopsin from Exiguobacterium sibiricum (ESR) and blue-light absorbing proteorhodopsin from the uncultured gamma-proteobacterium “Hot 75m4” (BPR).
Residues shared between all the protein variants are marked with asterisks. Single amino acid residue at position 106 (BPR numbering) that functions as a spectral tuning switch and accounts for most of the spectral difference between the two pigment families is highlighted in light grey. Primary proton acceptor and donor are highlighted in dark grey (D86) and with a frame (K97), respectively. The Schiff base (K232 for ESR, K226 for E17R) is indicated by a diamond. Residues differing between both green-PRs are indicated with arrows. The seven transmembrane α-helices are indicated with blue bubbles.