Fig. 2. Analysis of nucleotide exchange and hydrolysis by representative Gα subunits indicates that fast nucleotide exchange is an ancestral property.
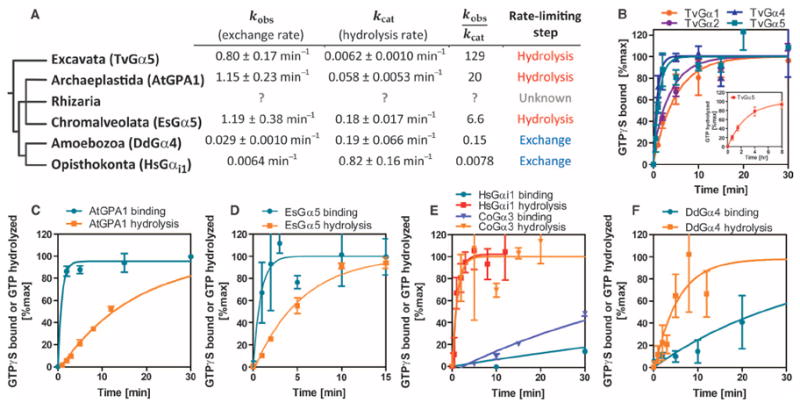
(A) Phylogeny showing the rates of nucleotide exchange and GTP hydrolysis of representative Gα subunits from each of the eukaryotic supergroups: T. vaginalis (Excavata), D. discoideum (Amoebozoa), C. owczarzaki (non-animal Opisthokonta), H. sapiens (Opisthokonta), and A. thaliana (Archae-plastida). See Fig. 1 and figs. S1 to S4 for the genes encoding GPCRs and 7TM-RGS proteins that are found in each clade. No information is available for Rhizaria because the genome was only recently released. Note that the origin of the nucleotide exchange–limited G cycle appears to have come after the split between the Amoebozoa and Opisthokonta and the Rhizaria, Chromalveolata, and Archaeplastida. The rates ± SE are computed from more than 12 data points shown in (B) to (F). A cutoff value of 1.0 in the kobs/kcat indicates the rate-limiting step. (B to F) Superimposed time courses of [35S]GTPγS binding and single-turnover [γ-32P]GTP hydrolysis in room temperature reactions containing 500 nM Gα proteins from C. owczarzaki, D. discoideum, A. thaliana, E. siliculosus, and H. sapiens. The [γ-32P]GTP hydrolysis data from the H. sapiens Gα protein are presented as means ± SEM of four independent experiments. The [35S]GTPγS binding data for T. vaginalis Gα proteins are presented as means ± SEM of three (TvGα1, TvGα2, and TvGα4) or seven (TvGα5) independent experiments. Nucleotide exchange and hydrolysis data for EsGα5, HsGαi1, AtGPA1, and CoGα3 and hydrolysis data for TvGα5 are calculated from means of at least two replicates, and the variation is expressed as SD.
