Figure 1.
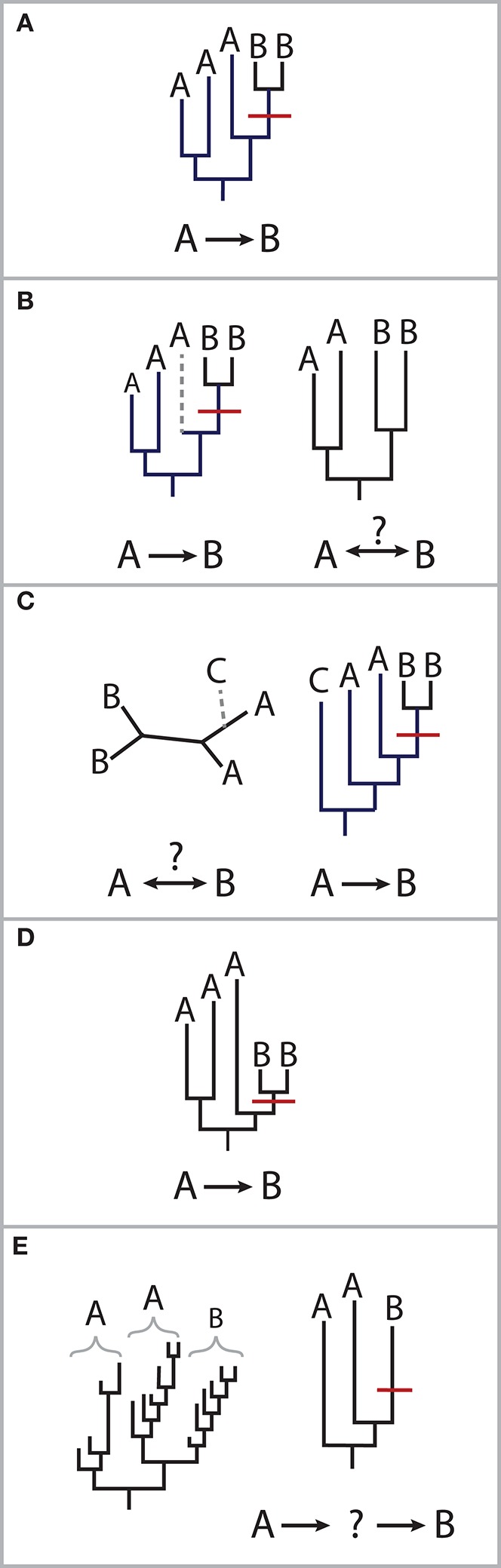
Scenarios for molecular epidemiology approaches. (A) Nesting of one individuals' strain lineages within another's supports transmission from the host carrying the ancestral strain to the host carrying the more recently diverged strain, as shown here of a putative transmission event (shown in red) from person A to person B. (B) The loss of lineages can affect our ability to determine directionality. Given the same phylogeny in (A), without the gray lineages, it is unclear which person's strains are ancestral. This can occur due to the choice of gene or characterizing fewer strains in an individual than what is present. (C) An outgroup helps distinguish transmission direction. Without lineage (C), it is unclear whether (A) transmitted strains to (B) or vice versa. The inclusion of appropriate control samples can help reduce the likelihood of indirect transmission from an intermediate host or environmental source. In the 1994 case involving HIV, controls were chosen from HIV-infected individuals in the same geography, although not necessarily with the same risk factors (i.e., drug use, sexuality, hemophilia; Metzker et al., 2002). (D) Phylogenetic distances may not reflect the timing of transmission. An organism's rate of evolution may depend on factors specific to the individual, such as immunity, diet or genetics, which create different host selective pressures. (E) The rate of evolution of the marker gene is important to detect putative direct transmission. Long-term carriage of a microbe with high rates of evolution may result in long branch-lengths, upon which it becomes more difficult to exclude the possibility of indirect transmission.
