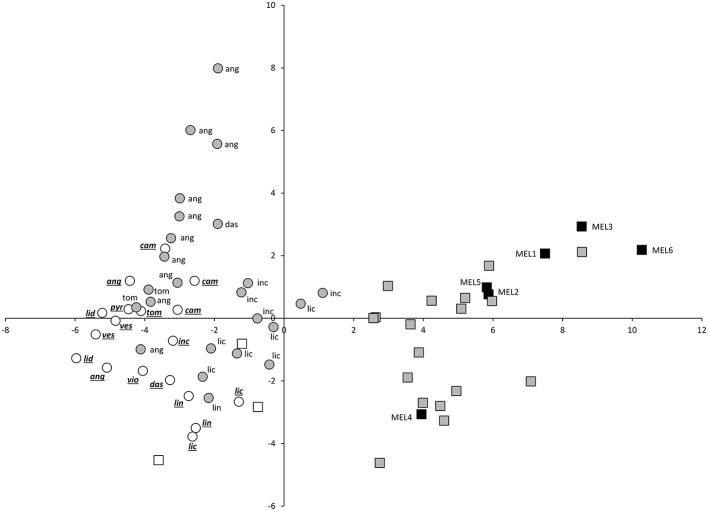
Figure 2.
First (X-axis) and second (Y-axis) principal components (37.2 and 12.0% of the total variation explained, respectively) scatterplot of cultivated eggplant, wild relatives and interspecific hybrids based on 27 conventional and 20 Tomato Analyzer morphological descriptors. Cultivated eggplant (S. melongena) is represented by black squares and the “MEL” accession code, primary genepool species S. insanum by white squares, interspecific hybrids between eggplant and S. insanum by gray squares, secondary genepool species by white circles (with species codes in underlined italics), and interspecific hybrids between eggplant and secondary genepool species by gray circles (with wild species codes in normal font). For secondary genepool species and their hybrids with eggplant, the following codes are used: ang (S. anguivi), cam (S. campylacanthum), das (S. dasyphyllum), inc (S. incanum), lic (S. lichtensteinii), lid (S. lidii), lin (S. linnaeanum), pyr (S. pyracanthos), tom (S. tomentosum), ves (S. vespertilio), vio (S. violaceum).