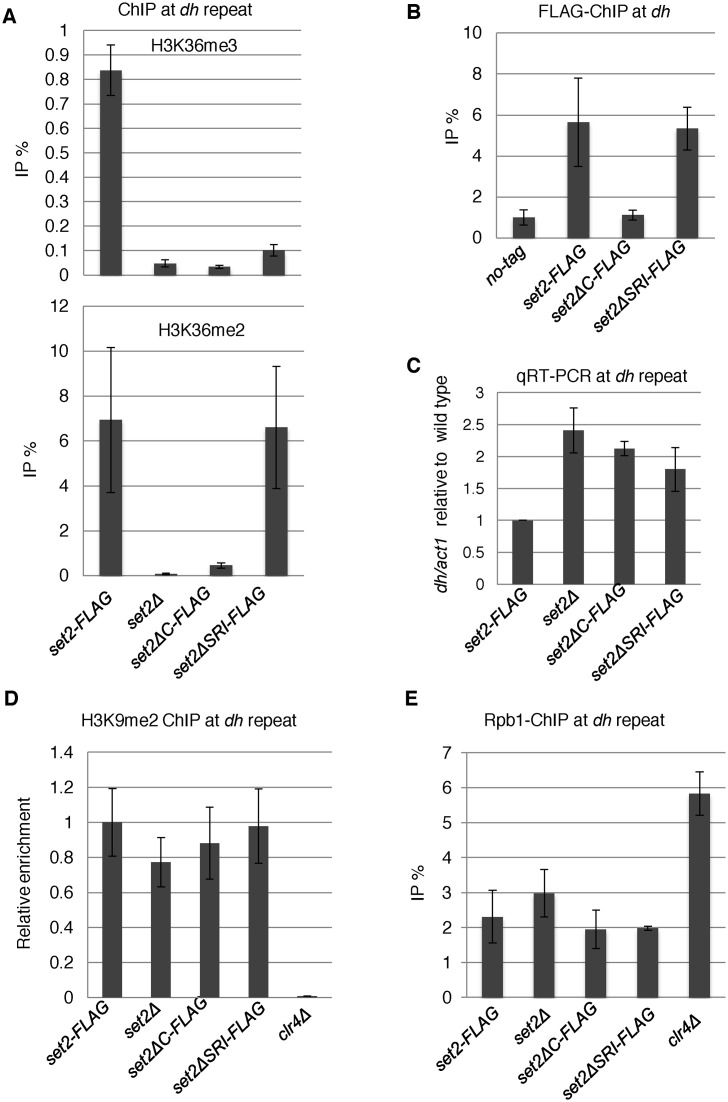
Figure 4.
Loss of Set2 compromised heterochromatic silencing without affecting heterochromatin structure. (A, B and D) ChIP analyses of H3K36me2 and H3K36me3 (A), Set2-FLAG (B), H3K9me2 (D) and Rpb1 (E) at heterochromatic dh repeats were performed using the indicated strains. Error bars show the standard deviation of three independent experiments. (C) Quantitative RT-PCR (qRT-PCR) analysis of the forward transcript from dh repeats was performed using the indicated strains. Error bars show the standard deviation of three independent experiments.