Figure 5.
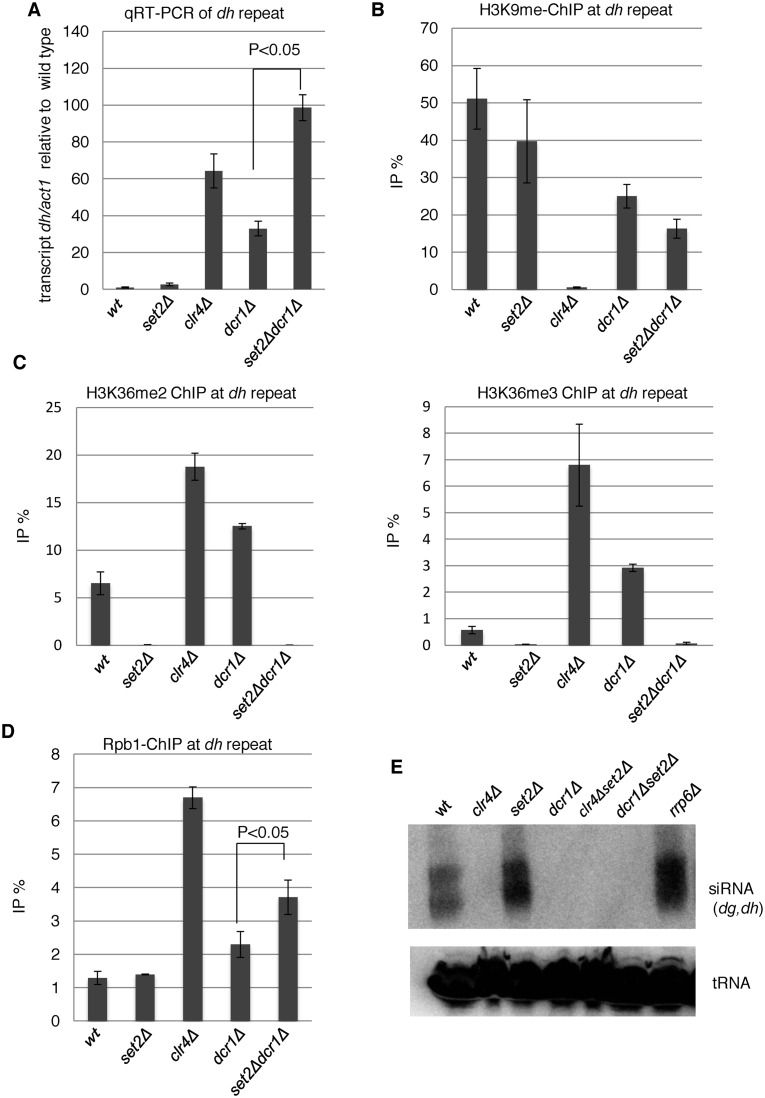
Set2 functions at heterochromatin independently of RNAi. (A) qRT-PCR analyses of the forward transcripts from dh repeats were performed using the indicated strains. Error bars show the standard deviation of three independent experiments. P-values were determined using a two-sided Student's t-test to compare dcr1Δset2Δ cells with dcr1Δ cells. (B and D) ChIP analyses of H3K9me2 (B) and Rpb1 (D) at heterochromatic dh repeats were performed using the indicated strains. Error bars show the standard deviation of three independent experiments. P-values were determined using a two-sided Student's t-test to compare dcr1Δset2Δ cells with dcr1Δ cells. (C) H3K36 methylation was increased in mutants that compromised heterochromatin. ChIP analyses of H3K36me2 (left panel) and H3K36me3 (right panel) at heterochromatic dh repeats were performed in the indicated strains. Error bars show the standard deviation of three independent experiments. (E) siRNA analysis by northern blotting using oligonucleotide probes specific for dg and dh centromeric repeats (40). Oligonucleotide probe specific for tRNAAsn was used as a loading control.