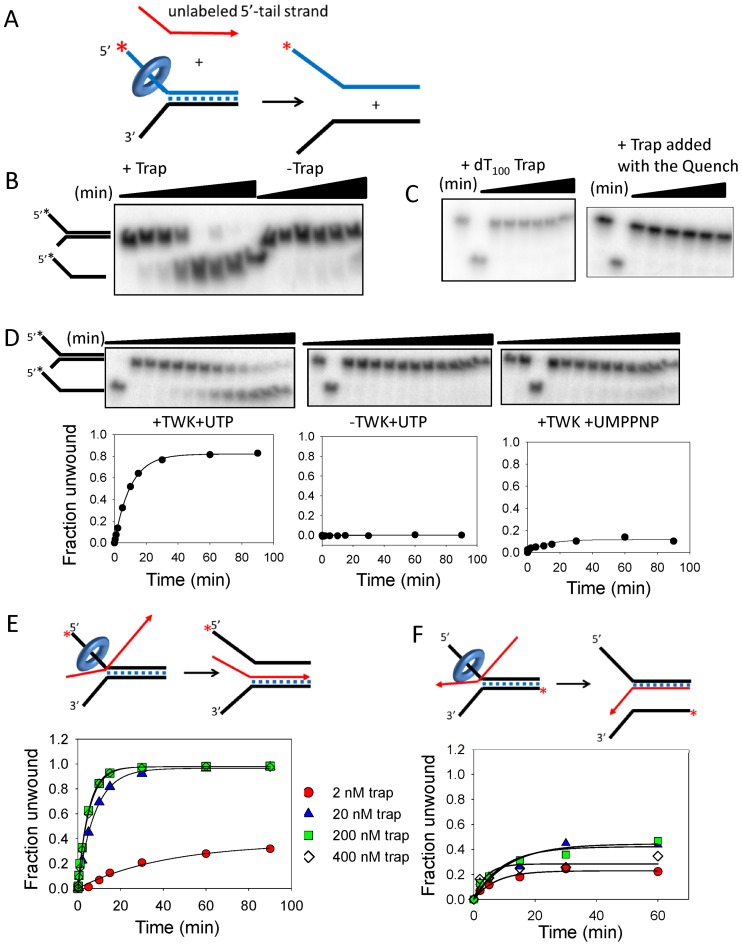
Figure 1.
DNA unwinding activity of the TWINKLE depends on trapping of the unwound strand by complementary ssDNA. (A) Twinkle was incubated with the fork DNA with UTP minus Mg(II) and reactions were initiated with Mg(II) plus and minus trap DNA. (B) Gel image shows the time course of 40-bp fork DNA strand separation in the presence and absence of 20 nM ssDNA trap (unlabeled 5′-tail strand, displaced-strand trap). (C) Left, DNA unwinding with dT100 trap (0–60 min) and right, DNA unwinding with displaced-strand trap (200 nM) added with the quenching solution. (D) Top, gel images shows the time course of DNA unwinding in the presence of 20 nM trap with Twinkle plus UTP (left), minus Twinkle (middle) and Twinkle plus UMPPNP (right). Bottom, quantitation of the gel images shows 5′-tail strand generation with an initial rate (exponential rate × amplitude) of 0.08 strand/min. (E) The unwinding efficiency depends on the trap concentration. The initial rates of DNA unwinding with displaced-strand trap at 2 nM (0.009 ± 0.002 strand/min), 20 nM (0.1 ± 0.01 strand/min), 200 nM (0.2 ± 0.006 strand/min) and 400 nM (0.19 ± 0.004 strand/min) is obtained from the kinetics. (F) The initial rates of DNA unwinding with unlabeled 3′-tail strand at 2 nM (0.03 ± 0.006 strand/min), 20 nM (0.04 ± 0.013 strand/min), 200 nM (0.04 ± 0.013 strand/min) and 400 nM (0.07 ± 0.03 strand/min) is obtained from the kinetics. The cartoons show Twinkle (ring) tracking on the 5′-tail strand of the fork DNA to displace the 3′-tail strand and simultaneously annealing either the 3′-tail strand to unlabeled displaced-strand trap (left, 1E), or the 5′-tail strand to the unlabeled 3′-tail strand (right, 1F). The standard errors (SE) of the initial rates are calculated from the SE of the exponential rates and amplitudes using the error propagation formula.